What We’re Reading: July 28th
 This week’s edition of What We’re Reading is guest edited by Dr. Bhavisha P. Sheth, who is currently working as a DST- Science, Technology and Innovation Policy Postdoctoral Fellow at the Entrepreneurship Development Institute of India (EDII), India. Her current research work focusses on Policy research studies of Science and Technology Entrepreneurship. She pursued a PhD in Plant Biotechnology and her thesis was awarded ‘the Best Life Sciences PhD Thesis’ in the state. During her PhD, she was a recipient of the UGC Junior and Senior Research Fellowships from Govt. of India. She has published 15 papers in national and international journals of high repute as well as presented her research in various conferences in India and abroad.
This week’s edition of What We’re Reading is guest edited by Dr. Bhavisha P. Sheth, who is currently working as a DST- Science, Technology and Innovation Policy Postdoctoral Fellow at the Entrepreneurship Development Institute of India (EDII), India. Her current research work focusses on Policy research studies of Science and Technology Entrepreneurship. She pursued a PhD in Plant Biotechnology and her thesis was awarded ‘the Best Life Sciences PhD Thesis’ in the state. During her PhD, she was a recipient of the UGC Junior and Senior Research Fellowships from Govt. of India. She has published 15 papers in national and international journals of high repute as well as presented her research in various conferences in India and abroad.
Differentially regulated orthologs in sorghum and the subgenomes of maize
 Gene expression patterns are often used to assign function, but how tight is the correlation between expression and function? Zhang et al. addressed this question by investigating whether orthologous genes in two species (including the two subgenomes of maize) show similar patterns of expression. Specifically, they carried out a genome-wide transcriptional comparison of cold-response in sorghum and maize. They found that most orthologous genes were not similarly regulated in maize and sorghum, although early-expressed cold-response genes showed greater cross-species conservation than later genes. Further, genes with conserved transcriptional regulation patterns had a low ratio of non-synonymous to synonymous substitutions, suggesting stronger selection. The authors suggest that cross-species expression comparisons can reveal functionally important genes. Plant Cell.
Gene expression patterns are often used to assign function, but how tight is the correlation between expression and function? Zhang et al. addressed this question by investigating whether orthologous genes in two species (including the two subgenomes of maize) show similar patterns of expression. Specifically, they carried out a genome-wide transcriptional comparison of cold-response in sorghum and maize. They found that most orthologous genes were not similarly regulated in maize and sorghum, although early-expressed cold-response genes showed greater cross-species conservation than later genes. Further, genes with conserved transcriptional regulation patterns had a low ratio of non-synonymous to synonymous substitutions, suggesting stronger selection. The authors suggest that cross-species expression comparisons can reveal functionally important genes. Plant Cell.
A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone ($)
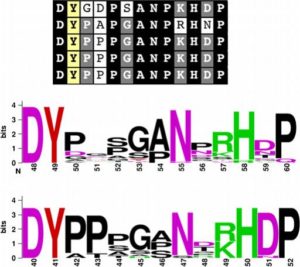 By measuring the length of Arabidopsis and rice roots treated with synthetic peptides, Pruitt et al. showed that RaxX, a peptide produced by the rice pathogen Xanthomonas oryzae pv. oryzae (Xoo), mimicked the growth-stimulating activity of plant peptides containing sulfated tyrosine residues (PSYs). They propose that the pathogen produces RaxX to promote growth of infected tissues, and they also show that RaxX-deficient Xanthomonas have decreased virulence. Unlike growth-promoting PSYs, the pathogen-derived peptide induces immune responses, indicating that the plant is able to distinguish between the endogenous and pathogen-derived peptides. New Phytol. 10.1111/nph.14609
By measuring the length of Arabidopsis and rice roots treated with synthetic peptides, Pruitt et al. showed that RaxX, a peptide produced by the rice pathogen Xanthomonas oryzae pv. oryzae (Xoo), mimicked the growth-stimulating activity of plant peptides containing sulfated tyrosine residues (PSYs). They propose that the pathogen produces RaxX to promote growth of infected tissues, and they also show that RaxX-deficient Xanthomonas have decreased virulence. Unlike growth-promoting PSYs, the pathogen-derived peptide induces immune responses, indicating that the plant is able to distinguish between the endogenous and pathogen-derived peptides. New Phytol. 10.1111/nph.14609
Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence ($)
 Ribeiro et al. describe functional aspects of the rice gene OsAPX4 encoding a peroxisomal ascorbate peroxidase, which shows the highest level of expression in leaves. RNAi silencing of this gene did not lead to an increase in the intracellular H2O2 levels and no change was observed in growing plants. However, a drastic change was seen in aging plants which exhibited early senescence. The study suggests that OsAPX4 plays a significant role in ROS signalling during leaf senescence Plant Science. 10.1016/j.plantsci.2017.07.009
Ribeiro et al. describe functional aspects of the rice gene OsAPX4 encoding a peroxisomal ascorbate peroxidase, which shows the highest level of expression in leaves. RNAi silencing of this gene did not lead to an increase in the intracellular H2O2 levels and no change was observed in growing plants. However, a drastic change was seen in aging plants which exhibited early senescence. The study suggests that OsAPX4 plays a significant role in ROS signalling during leaf senescence Plant Science. 10.1016/j.plantsci.2017.07.009
Heterologous expression of two GPATs from Jatropha curcas alters seed oil levels in transgenic Arabidopsis thaliana ($)
 Mishra et al. (2017) characterized two GPATs (glycerol-3-phosphate acyltransferase) JcGPAT1 and JcGPAT2 in the biofuel seed oil crop Jatropha curcas. These GPATs showed sequence homology with Arabidopsis acyltransferase-1 (ATS1) and Arabidopsis GPAT (AtGPAT9) respectively. Subcellular localization studies indicated that JcGPAT1 is localized in plastids whereas JcGPAT2 is localized in the endoplasmic reticulum. Overexpression of these enzymes in Arabidopsis thaliana led to an increase in total seed oil content – as much as a 43-60% increase in transgenic lines overexpressing JcGPAT2. This study highlights the role of these GPATs in oil biosynthesis in Jatropha curcas. Plant Science 10.1016/j.plantsci.2017.07.003 Image source: Elitre
Mishra et al. (2017) characterized two GPATs (glycerol-3-phosphate acyltransferase) JcGPAT1 and JcGPAT2 in the biofuel seed oil crop Jatropha curcas. These GPATs showed sequence homology with Arabidopsis acyltransferase-1 (ATS1) and Arabidopsis GPAT (AtGPAT9) respectively. Subcellular localization studies indicated that JcGPAT1 is localized in plastids whereas JcGPAT2 is localized in the endoplasmic reticulum. Overexpression of these enzymes in Arabidopsis thaliana led to an increase in total seed oil content – as much as a 43-60% increase in transgenic lines overexpressing JcGPAT2. This study highlights the role of these GPATs in oil biosynthesis in Jatropha curcas. Plant Science 10.1016/j.plantsci.2017.07.003 Image source: Elitre
Phylogenetic analysis of proteins involved in the stringent response in plant cells ($)
 The stringent response in bacteria is a global transcriptional response to stress, mediated by the second messenger ppGpp (guanosine 5′-diphosphate 3′-diphosphate) which is synthesized by the action of guanosine 5′-triphosphate 3′-diphosphate pyrophosphatase (GppA) or exopolyphosphatase (Ppx). Recently it was shown that plants also exhibit a stringent response mediated by ppGpp, but the origin of this response in plants has not been resolved. Ito et al. investigated the distribution of the gppA/ppx gene families as well as the RSH hydrolase gene family. Their data indicate that genes involved in the stringent response were likely introduced to plants from various bacterial phyla by lateral gene transfer events. Moreover, the results suggest that plant RSH homologs likely function in the plastid, whereas the plant gppA/ppx homologs may function in the cytosol. J. Plant Research 10.1007/s10265-017-0922-8
The stringent response in bacteria is a global transcriptional response to stress, mediated by the second messenger ppGpp (guanosine 5′-diphosphate 3′-diphosphate) which is synthesized by the action of guanosine 5′-triphosphate 3′-diphosphate pyrophosphatase (GppA) or exopolyphosphatase (Ppx). Recently it was shown that plants also exhibit a stringent response mediated by ppGpp, but the origin of this response in plants has not been resolved. Ito et al. investigated the distribution of the gppA/ppx gene families as well as the RSH hydrolase gene family. Their data indicate that genes involved in the stringent response were likely introduced to plants from various bacterial phyla by lateral gene transfer events. Moreover, the results suggest that plant RSH homologs likely function in the plastid, whereas the plant gppA/ppx homologs may function in the cytosol. J. Plant Research 10.1007/s10265-017-0922-8
Structure and expression patterns of dehydrin gene family in barley ($)
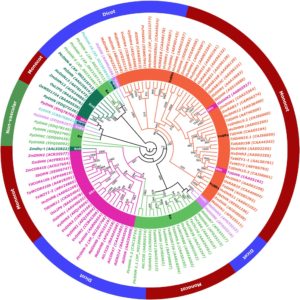 Dehydrins are proteins that have important roles in dehydration stress in plants. Abedini et al. carried out a phylogenetic analysis of barley dehydrins and investigated physiological and gene expression levels in tolerant (HV1), and susceptible (HV2) cultivars and a drought tolerant Iranian wild barley genotype (Spontaneum; HS) subjected to water stress and after recovery. They found notable differences in physiological responses and dehydrin expression levels in the drought tolerant varieties as compared to the susceptible variety, as well as between the wild and cultivated genotypes, supporting the role for dehydrins in vegetative drought tolerance. As barley is an important grain in arid regions, the study will contribute to future breeding efforts for drought tolerance J. Plant Research 10.1007/s10265-017-0941-5.
Dehydrins are proteins that have important roles in dehydration stress in plants. Abedini et al. carried out a phylogenetic analysis of barley dehydrins and investigated physiological and gene expression levels in tolerant (HV1), and susceptible (HV2) cultivars and a drought tolerant Iranian wild barley genotype (Spontaneum; HS) subjected to water stress and after recovery. They found notable differences in physiological responses and dehydrin expression levels in the drought tolerant varieties as compared to the susceptible variety, as well as between the wild and cultivated genotypes, supporting the role for dehydrins in vegetative drought tolerance. As barley is an important grain in arid regions, the study will contribute to future breeding efforts for drought tolerance J. Plant Research 10.1007/s10265-017-0941-5.
Identification and functional analysis of new peroxygenases in oat ($)
 Peroxygenases catalyze the hydroperoxide dependent epoxidation of unsaturated fatty acids. Benaragama et al. identified two novel peroxygenases (AsPXG2 and AsPXG3) in oat (Avena sativa L.) AsPXG2 and AsPXG3 share structural similarity by having a single transmembrane domain, conserved histidines for heme-binding as well as a conserved EF-hand motif. However, they show only 50% amino acid identity with each other, and the activity of heterologously-expressed AsPXG3 is much higher than that of AsPXG2. Site-directed mutagenesis of AsPXG3 showed that replacement of two conserved histidines by alanine resulted in complete loss of activity, and substitution of three conserved residues surrounding the two histidines resulted in reduction of enzymatic activity by more than 80%, suggesting that these conserved residues are located near the catalytic sites, where they coordinate the heme group and define the shape and size of the catalytic site. Planta 10.1007/s00425-017-2729-1.
Peroxygenases catalyze the hydroperoxide dependent epoxidation of unsaturated fatty acids. Benaragama et al. identified two novel peroxygenases (AsPXG2 and AsPXG3) in oat (Avena sativa L.) AsPXG2 and AsPXG3 share structural similarity by having a single transmembrane domain, conserved histidines for heme-binding as well as a conserved EF-hand motif. However, they show only 50% amino acid identity with each other, and the activity of heterologously-expressed AsPXG3 is much higher than that of AsPXG2. Site-directed mutagenesis of AsPXG3 showed that replacement of two conserved histidines by alanine resulted in complete loss of activity, and substitution of three conserved residues surrounding the two histidines resulted in reduction of enzymatic activity by more than 80%, suggesting that these conserved residues are located near the catalytic sites, where they coordinate the heme group and define the shape and size of the catalytic site. Planta 10.1007/s00425-017-2729-1.
Vesicle dynamics during plant cell cytokinesis reveals distinct developmental phases
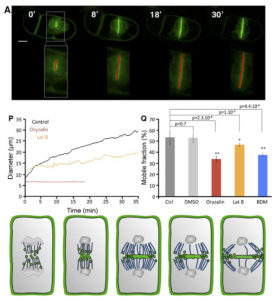 The cell plate is a common feature of plant cell divisions, but the mechanisms forming this structure are not completely understood. The cell plate is made from membrane vesicles carrying polysaccharide cargo, and thousands of tiny vesicles fuse together to form one cell plate. Observing fluorescence micrographs alone cannot provide a full picture of cell plate biogenesis, so Oostende-Triplet et al. developed FluMOS (Fluorescence Morphological Operators Software). This program can identify and analyze cell plates within fluorescence confocal images. The authors identified three distinct phases of cell plate expansion in N. tabacum BY-2 cells based on the growth rate of the cell plate. FluMOS quantitated vesicle movement towards nascent cell plates. Initially, the vesicles move quickly to the cell plate when its growth begins. As the cell plate growth, vesicle movement remains high at the edges, but vesicle movement slows in the center of the cell plate. Pharmacological inhibition of membrane trafficking and protein synthesis showed both processes contribute to cell plate expansion. (Summary by Daniel Czerny) Plant Physiol. 10.1104/pp.17.00343
The cell plate is a common feature of plant cell divisions, but the mechanisms forming this structure are not completely understood. The cell plate is made from membrane vesicles carrying polysaccharide cargo, and thousands of tiny vesicles fuse together to form one cell plate. Observing fluorescence micrographs alone cannot provide a full picture of cell plate biogenesis, so Oostende-Triplet et al. developed FluMOS (Fluorescence Morphological Operators Software). This program can identify and analyze cell plates within fluorescence confocal images. The authors identified three distinct phases of cell plate expansion in N. tabacum BY-2 cells based on the growth rate of the cell plate. FluMOS quantitated vesicle movement towards nascent cell plates. Initially, the vesicles move quickly to the cell plate when its growth begins. As the cell plate growth, vesicle movement remains high at the edges, but vesicle movement slows in the center of the cell plate. Pharmacological inhibition of membrane trafficking and protein synthesis showed both processes contribute to cell plate expansion. (Summary by Daniel Czerny) Plant Physiol. 10.1104/pp.17.00343
Are you interested in contributing to What We’re Reading? We welcome informal, one-off summaries, but you can also formalize your contributions by applying to be a Plantae Fellow, like Daniel and Bhavisha are!



Leave a Reply
Want to join the discussion?Feel free to contribute!