The same but different: CoMoVa, an algorithm to identify functional variation in cis regulatory elements
Transcription factors (TFs) act through TF binding sites (TFBSs) to control the transcription of associated genes. TFBSs are short and degenerate sequences that are often depicted using a Position Weight Matrix, which contain invariant nucleotides that are crucial for TF binding and variable nucleotides that minimally influence TF binding. The functionality of these variable nucleotides is poorly understood and the frequency with which TFBSs are distributed throughout the genome raises difficulties with the functional assignment of any given TFBS. The problems with connecting TF function with specific TFBSs is further exacerbated by large families of TFs in plants, where related TFs may differ in their degrees of specificity for a TFBS (Boer et al., 2015). The activities of the AUXIN RESPONSE FACTOR (ARF) TFs, which mediate the response to the phytohormone auxin in plants, exemplify many of these issues. In Arabidopsis thaliana, 22 genes encode ARF-TFs that bind to auxin response elements (AuxREs, TGTCNN, where N is an A, T, C, or G nucleotide) in the promoters of genes to control transcription (Finet et al., 2012). The identification of AuxREs facilitated the development of an auxin sensor (DR5), which is based on a transgene harboring eight repeats of an AuxRE variant (TGTCTC) that drives the expression of reporter protein (Ulmasov et al., 1997). This sensor was recently modified by changing the identity of the nucleotides in the variable region of the AuxRE (TGTCGG), which improved the strength of the auxin response (Liao et al., 2015). However, the relevance of these modifications to the biological functions of ARF-TFs was unclear.
 To test whether variable nucleotides within AuxREs were associated with distinct functions, Lieberman-Lazarovich et al. developed a novel algorithm to identify conserved AuxRE variants. The authors developed an algorithm, CoMoVa (Conservation of Motif Variants) that detects the conservation of TFBSs by estimating the background rates of nucleotide variation surrounding the TFBS. The authors applied this algorithm to 45 angiosperms to detect AuxREs with the sequence TGTCNN (where N is an A, T, C, or G nucleotide). CoMoVa detected conservation of specific AuxRE variants in over 200 genes across these angiosperms. Conservation was detected for several AuxRE variants, including AuxREs with CC or GG in the variable region. Gene ontology analysis showed that ‘Response to auxin’ was enriched for genes containing CC variants whereas genes with cell wall synthesis functions were more likely to contain GG variants.
To test whether variable nucleotides within AuxREs were associated with distinct functions, Lieberman-Lazarovich et al. developed a novel algorithm to identify conserved AuxRE variants. The authors developed an algorithm, CoMoVa (Conservation of Motif Variants) that detects the conservation of TFBSs by estimating the background rates of nucleotide variation surrounding the TFBS. The authors applied this algorithm to 45 angiosperms to detect AuxREs with the sequence TGTCNN (where N is an A, T, C, or G nucleotide). CoMoVa detected conservation of specific AuxRE variants in over 200 genes across these angiosperms. Conservation was detected for several AuxRE variants, including AuxREs with CC or GG in the variable region. Gene ontology analysis showed that ‘Response to auxin’ was enriched for genes containing CC variants whereas genes with cell wall synthesis functions were more likely to contain GG variants.
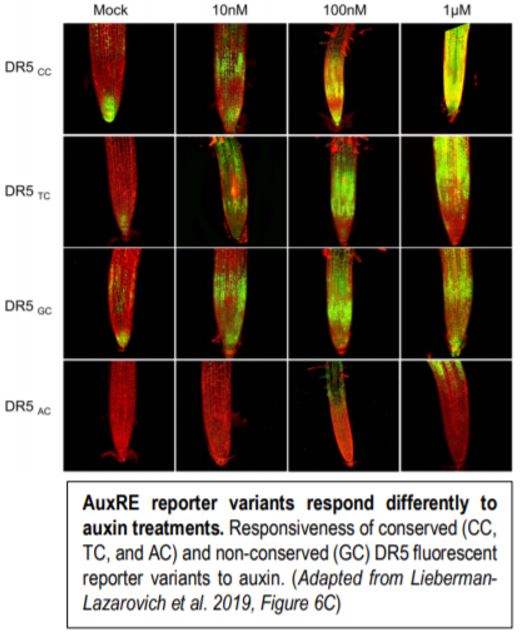
The authors then generated several auxin sensors based on the DR5 sensor design with the conserved variants AC, TC, CC, and the non-conserved variant GC. Each of these variants responded differently to auxin treatments with the CC variants being the most responsive and AC being the least responsive (Figure). The weak response of the AC variant to auxin was unexpected as 18 genes harbored this conserved motif in their regulatory regions and this motif was shown to interact with ARF-TFs in vitro (Boer et al., 2014). The authors demonstrated that although this AuxRE variant was less active, this could be offset by promoting higher levels of ARF accumulation. Furthermore, the expression pattern of the AC variant was restricted to the root elongation zone of plants treated with auxin whereas the other variants were more broadly expressed (Figure). The authors argue that the weak and tissue specific responsiveness the AC variant to auxin is important to elicit specific spatiotemporal transcriptional responses from associated genes. The authors then designed a novel auxin sensor based on the conserved position of the AuxRE motif to the transcriptional start site and the CC AuxRE variant, both of which were identified using CoMoVa. The expression pattern of this sensor was broader than the DR5 sensor in both Arabidopsis and tomato (Solanum lycopersicum), suggesting that this new sensor is more sensitive than the original DR5 sensor.
CoMoVa was also used identify conserved variants in the TFBSs that two other phytohormone responses (abscisic acid and cytokinin), some of which could also be functionally classified. Further implementation of CoMoVa and follow-up functional studies, such as the ones presented here, promise to improve our understanding of how variations in TFBSs lead to specific transcriptional responses.
Diarmuid S. Ó’Maoiléidigh
Institute of Integrative Biology
University of Liverpool,
dsom@liverpool.ac.uk
ORCID ID: 0000-0002-3043-3750
References
Boer, D., Freire-Rios, A., and van den Berg, W. (2014). Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors. Cell 156: 577–89. https://doi: 10.1016/j.cell.2013.12.027.
Finet, C., Berne-Dedieu, A., Scutt, C.P., and Marlétaz, F. (2012) Evolution of the ARF Gene Family in Land Plants: Old Domains, New Tricks. Mol. Biol. Evol. 30(1):45–56 https://doi:10.1093/molbev/mss220
Lieberman-Lazarovich, M., Yahav, C., Israeli, A., and Efroni, I. (2019) Deep Conservation of Cis-element Variants Regulating Plant Hormonal Responses. Plant Cell DOI: https://doi.org/10.1105/tpc.19.00129.
Liao, C.-Y., Smet, W., Brunoud, G., Yoshida, S., Vernoux, T., and Weijers, D. (2015). Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12: 207–210. https://doi.org/10.1038/nmeth.3279
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–71. https://doi.org/10.1105/tpc.9.11.1963