Pseudogenes in Plants: Their Associations with Long Noncoding RNAs and MicroRNAs
Xie et al. applied a multifaceted approach to study plant pseudogenes, combining RNA-seq, comparative genomics, and molecular biology. They conclude that rapid rewiring of psuedogene transcriptional regulatory regions is a major mechanism driving the origin of novel regulatory modules. Plant Cell https://doi.org/10.1105/tpc.18.00601
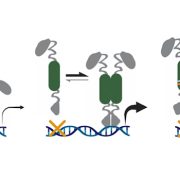
Background: Genomes contain a range of elements including not only genes, but also long noncoding RNAs (lncRNAs) and microRNAs (miRNAs). LncRNAs and miRNAs are now known to regulate genes. In addition, an organism’s genome contains so-called “genomic fossils”, such as pseudogenes. Pseudogenes are disabled copies of functional genes that have been retained in the genome. Pseudogenes have been thought to be non-functional, but previous studies have reported that some lncRNAs/miRNAs overlap with pseudogenes.
Question: We wanted to elucidate whether pseudogenes have any functions. We thus examined their relationships at genome-wide level. We developed a computational pipeline (PlantPseudo) that can search a plant genome and identify pseudogenes in the genome. We used this pipeline to clarify the relationships between pseudogenes and genes/lncRNAs/miRNAs on a genome-wide scale.
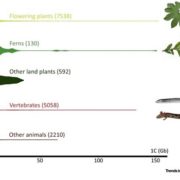
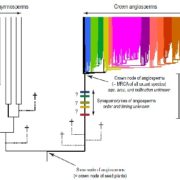
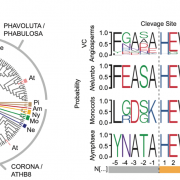
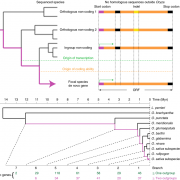
Findings: We identified approximately 250,000 pseudogenes in seven plant species: Arabidopsis thaliana, Brachypodium distachion, Glycine max, Medicago truncatula, Oryza sativa, Populus trichocarpa, and Sorghum bicolor. The larger genomes had more pseudogenes. Most of the pseudogenes were present only in one organism, suggesting their fast evolution. Studying the positions of pseudogenes showed that their distribution was not uniform along the genomes. Only a few were transcribed into RNA molecules. In addition, we found that a surprisingly large fraction of lncRNAs/miRNAs were distributed in the flanking regions of pseudogenes, indicating their origin is highly associated with pseudogenes.
Next steps: Our work supports the ongoing effort to study the mechanism that contributes to the origin of LncRNAs/miRNAs. Our long-term goal to is to make functional interpretations of the pseudogenes that shape plant evolution.
Jianbo Xie, Ying Li, Xiaomin Liu, Yiyang Zhao, Bailian Li, Pär K. Ingvarsson and Deqiang Zhang. (2019). Evolutionary Origins of Pseudogenes and Their Association with Regulatory Sequences in Plants. Plant Cell. https://doi.org/10.1105/tpc.18.00601.
Key words: pseudogenes, evolution, lncRNA, miRNA