Polyploid Pairing Problems: How Centromere Repeat Divergence Helps Wheat Sort It All Out
By Jennifer Mach, Science Editor
I don’t know how polyploids get their chromosomes paired correctly in meiosis— I can’t even sort my socks. Sometimes I give up and pair random socks, but plants can’t go with random pairings, because that’s a route to death, and with genomes in the dozens of Gb in tens of chromosomes, polyploid plants like Triticum aestivum (hexaploid, 17 Gb) have one heck of a sorting problem. How does each wheat chromosome find its one and only homolog and exclude the four homoeologs? Despite having several sets of very similar chromosomes, the chromosomes in polyploids pair like they do in diploids, and how they do this remains unclear. Pairing of chromosomes involves specific chromosome regions, including the centromere—that delightfully complex repetitive region that has frustrated genome sequencing efforts and functional analysis in so many species. The dynamics of these regions have been well studied; for example, studies in T. aestivum show that the centromeres of six homoeologs interact before homologs pair off (reviewed in da Innes and White, 2015), but the mechanisms remain unclear.
To explore the question of centromere sequence variation in polyploids, Su et al. (2019) examine the centromeres of wheat chromosomes, using data from chromatin immunoprecipitation (ChIP) with the centromere-specific histone H3 variant CENH3 to identify functional centromere regions. They mapped their ChIP-sequencing data from the hexaploid variety Chinese Spring (Guo et al., 2016) to the T. aestivum reference genome. Including the sequences that mapped to unincorporated scaffolds (typical of hard-to-align centromere sequences), they estimate that functional wheat centromeres are around 7.8 Mb.
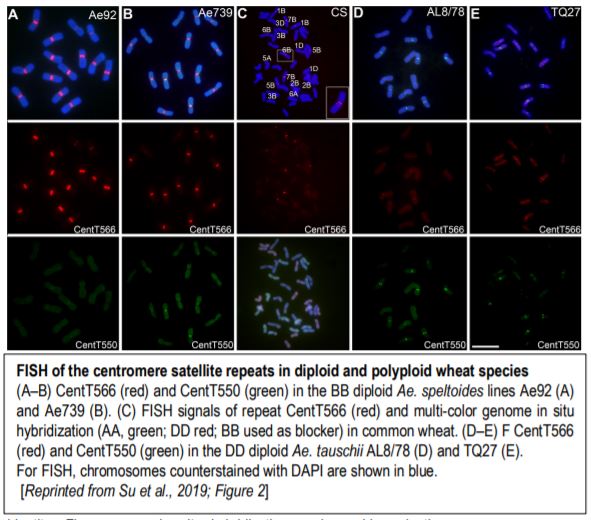 Plant centromeres generally contain many transposable elements and large arrays of tandem repeats of sequences in the 150–180 bp range, perfect for looping around a CENH3 nucleosome. Computational identification of repeats followed by comparison of repeat enrichment in the ChIP-seq data for CENH3 found centromere-specific long-terminal repeat retrotransposons (typical for grass species), and two tandem repeats, CenT566 (566 bp) and CenT550 (550 bp). These two repeats share little or no sequence identity. Fluorescence in situ hybridization (FISH) confirmed the centromere localization of both CenT repeats (figure). Analysis of nucleosome positioning indicated that CENH3 nucleosomes are phased on the CenT550 and CenT566 repeats, tending to center around WW (W=A or T) dinucleotides.
Plant centromeres generally contain many transposable elements and large arrays of tandem repeats of sequences in the 150–180 bp range, perfect for looping around a CENH3 nucleosome. Computational identification of repeats followed by comparison of repeat enrichment in the ChIP-seq data for CENH3 found centromere-specific long-terminal repeat retrotransposons (typical for grass species), and two tandem repeats, CenT566 (566 bp) and CenT550 (550 bp). These two repeats share little or no sequence identity. Fluorescence in situ hybridization (FISH) confirmed the centromere localization of both CenT repeats (figure). Analysis of nucleosome positioning indicated that CENH3 nucleosomes are phased on the CenT550 and CenT566 repeats, tending to center around WW (W=A or T) dinucleotides.
Although the actual progenitors of hexaploid wheat died thousands of years ago, their descendants Triticum urartu (A genome), Aegilops speltoides (B genome), and Aegilops tauschii (D genome) give us a good idea of the species that hybridized to make hexaploid wheat (AABBDD). Indeed, CenT566 occurred on all centromeres of Ae. speltoides and CenT550 occurred on several centromeres of Ae. tauschii but in the other diploids, these repeats only occurred on a few centromeres (figure). In hexaploid wheat, CenT566 occurred in all centromeres of the B genome and in a few A and D centromeres. Moreover, CenT550 was detected in several D centromeres and one B centromere. Some T. aestivum centromeres showed no FISH signal for either repeat and some showed expanded repeat domains.
To examine the variation in these repeats during polyploidization, the authors identified polymorphisms in the monomer sequences. CenT566 repeats clustered into three groups, which generally mapped to the three subgenomes, with the most-homogeneous cluster mapping to the B centromeres. Moreover, many CenT566 tandem arrays occur outside of CENH3 regions, particularly in the A and D centromeres, as indicated by cytological and sequencing data. This suggests that more-homogeneous arrays form the centromere cores. The B genome diploids showed higher sequence identity for CenT566, indicating that this sequence diverged on the A and D chromosomes after polyploid formation. Moreover, examination of colinear genes showed centromere rearrangements between Ae. tauschii and T. aestivum.
With rare exceptions (hello, B chromosomes), losing a chromosome in meiosis has severe consequences, unlike losing a sock. The identification of CENH3-associated wheat centromere repeats and analysis of centromere structure reveals intriguing changes in centromere composition during polyploidy—and, as long-read sequencing technology improves, our understanding of the centromere, will improve. The specification and maintenance of functional centromeres involves epigenetic mechanisms that interact with the underlying sequence, which has complicated efforts to infer the functions of various centromere components, but the study of centromeres in polyploid plants paves the way for understanding the role that sequence variation plays in the specificity of centromere pairing.
REFERENCES
Da Ines, O., and White, C.I. (2015) Centromere Associations in Meiotic Chromosome Pairing. Annu. Rev. Genet. 2015. 49:95–114 10.1146/annurev-genet-112414-055107
Guo, X., Su, H., Shi, Q., Fu, S., Wang, J., Zhang, X., Hu, Z., and Han, F. (2016). De Novo Centromere Formation and Centromeric Sequence Expansion in Wheat and its Wide Hybrids. PLoS Genet. 12, e1005997.
Su, H., Liu, Y., Liu, C., Shi, Q., Huang, Y., and Han, F. (2019). Centromere Satellite Repeats Have Undergone Rapid Changes in Polyploid Wheat Subgenomes. Plant Cell DOI: https://doi.org/10.1105/tpc.19.00133.