Mapping the landscape of C4 gene regulation
Burgess et al characterize genome-wide patterns of transcription factor binding to provide insight into the architecture associated with C4 photosynthesis gene expression. Plant Cell https://doi.org/10.1105/tpc.19.00078
By Steven James Burgess (University of Cambridge, UK) and Ivan A. Reyna-Llorens (University of Cambridge, UK)
Background: Photosynthesis comes in various shapes and forms. In its most efficient incarnation, carbon fixation is split between mesophyll and bundle sheath cells in order to avoid the energetic penalties caused by photorespiration which competes with the Calvin-Benson Cycle. The C4 pathway has evolved in over 60 lineages of plants and provides a growth advantage in warm dry environments, such as savannah grasslands which are dominated by C4 species. While the biochemical reactions of C4 photosynthesis were elucidated over 50 years ago, we know relatively little about the genetic regulation required to compartment photosynthesis between two cell types.
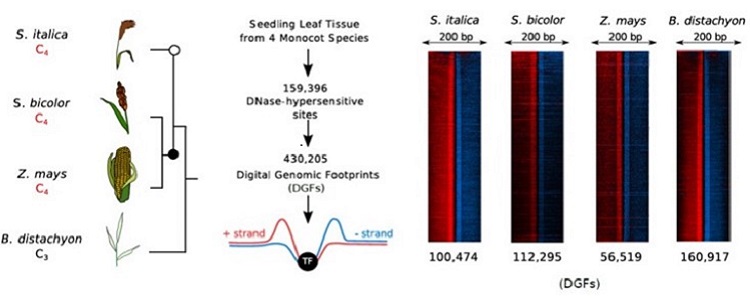
Question: We wanted to understand if the repeated evolution of mechanisms controlling the expression of C4 enzymes in a leaf could be explained by the use of the same DNA-regulatory elements. To do this we used a technique known as DNaseI-SEQ to map open chromatin, or regions of the genome accessible for transcription factors to bind, as well as individual DNA-elements bound by transcription factors in whole leaves and bundle sheath strands. We chose three C4 grasses, maize, sorghum and setaria and one non-C4 grass, Brachypodium to see if regulation was similar in two independent lineages of C4 photosynthesis.
Findings: We found that the whole leaf and bundle sheath generally share the same patterns of open chromatin, yet specific transcription factor binding sites are highly variable. Within a species, multiple C4 enzymes shared the same DNA-regulatory motifs. This opens up the possibility that the division of metabolism between mesophyll and bundle sheath cells in a C4 leaf is controlled by a relatively small number of transcription factors. In addition, by comparing between species, we found that few DNA-regulatory elements were shared by the same C4 enzymes. This suggests that regulation of C4 photosynthesis genes evolved in many ways, using a range of different DNA-regulatory elements and transcription factors.
Next Steps: This work identified a large number of regulatory elements that are bound by transcription factors in C4 and C3 leaves. The next steps include identifying the combination of these regulatory elements that are sufficient to control C4 gene expression, and as many transcription factors can bind to similar DNA-regulatory elements, defining the exact transcription factors binding to these elements. We also hope this will be a good resource to the community to explore the DNA binding landscape for their own genes of interest.
Steven J. Burgess, Ivan Reyna-Llorens, Sean R. Stevenson, Pallavi Singh, Katja Jaeger and Julian M. Hibberd. (2019). Genome-wide Transcription Factor Binding in Leaves from C3 and C4 Grasses. Plant Cell. https://doi.org/10.1105/tpc.19.00078.
Key words: Photosynthesis, evolution, gene regulation