Loss-of-Function, a Strategy for Adaptation in Arabidopsis
When thinking about adaptive processes, the concept of genetic innovation comes to mind. At the genomic scale, a lot of attention has been given to gene introgression, duplication, and novel functionalization to uncover the bases of adaptation. However, as the majority of genes are non-essential, both the gain and loss of genetic information can help plants adapt to their changing environments. The benefits of single gene losses are well documented in plants, for example, for variations in phytochemicals or transitions in pollination syndromes and selfing (Albat and Canestro, 2016). However, systematic analyses of loss-of-function mutations at the genome-wide level and their consequences at the global scale are lacking.
 Xu et al. (2019) performed a systematic analysis of a large consolidated genomic dataset of Arabidopsis thaliana sampled over Eurasia and Africa. They found that two-thirds of the protein-coding genes have loss-of-function variants in the over 1,000 wild populations analyzed. They also estimated that on average, 3% of the protein-coding genes present loss-of-function mutations in every wild population. These loss-of-function mutations are not present at random in the genome. Their frequency depends on nucleotide diversity and transposable element density. In addition, genetic redundancy is affected differentially, as loss-of-function variants correlate with gene family size.
Xu et al. (2019) performed a systematic analysis of a large consolidated genomic dataset of Arabidopsis thaliana sampled over Eurasia and Africa. They found that two-thirds of the protein-coding genes have loss-of-function variants in the over 1,000 wild populations analyzed. They also estimated that on average, 3% of the protein-coding genes present loss-of-function mutations in every wild population. These loss-of-function mutations are not present at random in the genome. Their frequency depends on nucleotide diversity and transposable element density. In addition, genetic redundancy is affected differentially, as loss-of-function variants correlate with gene family size.
The authors also estimated the environmental impact of these loss-of-function variants. They performed a genome-wide association study of the loss-of-function variants and variables in the WORDCLIM database. This analysis revealed 125 loss-of-function variants associated with climatic variables. This further confirms the findings of a recent study based on the 1001 Arabidopsis dataset that described the role of loss-of-function variants in drought tolerance strategies associated with flowering time (Monroe et al., 2018). These two publications show that plants use natural gene knockouts to adapt to climatic changes and environmental challenges. Furthermore, both studies found evidence of positive selection that maintains the resulting phenotypes.
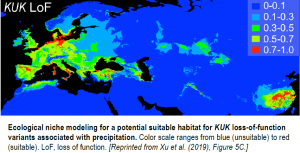
To further illustrate the power of loss-of-function mutations in shaping phenotypic variation, Xu et al. performed in-depth analysis of stop-gain mutations in KUK (KURZ UND KLEIN), PRR5 (PSEUDO-RESPONSE REGULATOR5), and LAZY1. KUK, a gene known to regulate root development, shows frequent loss-of-function variation across the wild populations and is associated with precipitation (see figure). The authors show that KUK stop-gain variants increase the length of the root meristem and mature cortical cell size. Stop-gain mutations in PRR5 were shown to slightly delay flowering time, while LAZY1, a rare loss-of-function variant, severely affects branch angles. Furthermore, in a Yangtze River basin population, the authors found that 1% of loss-of-function mutations were under positive selection, including KUK.
Xu et al. revealed the extent and potential of loss-of-function mutations in shaping the phenotypic diversity in Arabidopsis, a self-fertile species that maintains homozygous loss-of-function mutations at high frequency. Systematic studies in other plant species are needed to address the full extent of the roles of loss-of-function mutations in plant adaptation and how they are maintained both locally and globally. Furthermore, 99% of loss-of-function variants in Arabidopsis showed no association to climatic variables in the genome-wide association study, providing room for testing other aspects of plant fitness and adaptation. Further research also needs to address the functional mechanisms from loss-of-function mutations to phenotypes.
REFERENCES
Albalat, R. and Cañestro, C. (2016). Evolution by gene loss. Nature Reviews Genetics, 17(7), 379-391.
Monroe, J.G., Powell, T., Price, N., Mullen, J.L., Howard, A., Evans, K., Lovell, J.T., McKay J.K. (2018). Drought adaptation in Arabidopsis thaliana by extensive genetic loss-of-function. eLife, 7: e41038.
Xu, Y.C., Niu, X.M., Li. X.X., He, W., Chen, J..F, Zou, Y.P., Wu, Q., Zhang, Y.E., Busch, W., and Guo, Y.L. (2019) Adaptation and phenotypic diversification in Arabidopsis through loss-of-function mutations protein-coding genes. The Plant Cell. DOI: https://doi.org/10.1105/tpc.18.00791



