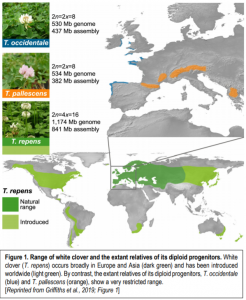
Looking Over Allopolyploid Clover
The allotetraploid species white clover (Trifolium repens) resulted from hybridization of two diploid European species whose extant relatives are found only in limited regions–– T. occidentale is a creeping clover found only in saline areas near the shores of Western Europe and T. pallescens is found only at restricted altitudes in Alpine habitats (figure). So, how did I come to be looking over a lovely patch of white clover in my lawn here in Chicago? Indeed, this legume serves as a valuable forage that grows nearly everywhere, with its native range extending across Western Asia, Europe, and Northern Africa and its introduced range extending around the world (figure).
When did the allopolyploidy event that produced T. repens occur, what were the genomic and transcriptomic consequences, and did this help white clover with its remarkable niche expansion? To examine the effects of allopolyploidy in clover, Griffiths et al. (2019) sequenced the genomes and transcriptomes (leaves, roots, shoots, and flowers) of T. repens, T. occidentale, and T. pallescens. Comparison of the full chloroplast genomes confirmed that T. pallescens represents the maternal parent and chloroplast donor for T. repens. Moreover, comparison of the ribosomal DNA in the nuclear genome confirmed that T. occidentale represents the other parent and the authors identified subgenome-specific polymorphisms in T. repens that corresponded to polymorphisms in the two diploids.
 Addressing the question of when, examination of the divergence of the three species, based on differences between pairwise alignments of the progenitor genomes themselves and the corresponding subgenomes, led to an estimated time of about 15–28,000 years ago, during the depths of the last glaciation. The authors speculate that the progenitors may have been growing together in glacial refugia—and indeed, the progenitors must have been together for some time because white clover appears to result from multiple hybridization events. Examination of re-sequencing data from four outbred individuals and genotyping-by-sequencing in white clover populations showed that T. repens did not undergo a major genetic bottleneck, as would be expected if it resulted from a single hybridization and chromosome doubling or fusion of unreduced gametes that produced only one progenitor. Rather, the data suggest that multiple hybridization events occurred in the early history of white clover. The authors speculate that this allowed a wider sampling of the genetic diversity of the progenitors, which was carried over into the emerging new species, underpinning the variation found in modern T. repens. Furthermore, white clover appears to have undergone a substantial population increase, consistent with it expanding to different niches after the glaciation.
Addressing the question of when, examination of the divergence of the three species, based on differences between pairwise alignments of the progenitor genomes themselves and the corresponding subgenomes, led to an estimated time of about 15–28,000 years ago, during the depths of the last glaciation. The authors speculate that the progenitors may have been growing together in glacial refugia—and indeed, the progenitors must have been together for some time because white clover appears to result from multiple hybridization events. Examination of re-sequencing data from four outbred individuals and genotyping-by-sequencing in white clover populations showed that T. repens did not undergo a major genetic bottleneck, as would be expected if it resulted from a single hybridization and chromosome doubling or fusion of unreduced gametes that produced only one progenitor. Rather, the data suggest that multiple hybridization events occurred in the early history of white clover. The authors speculate that this allowed a wider sampling of the genetic diversity of the progenitors, which was carried over into the emerging new species, underpinning the variation found in modern T. repens. Furthermore, white clover appears to have undergone a substantial population increase, consistent with it expanding to different niches after the glaciation.
Addressing the question of the genomic and transcriptomic effects of allopolyploidy, the authors find that white clover has retained most of the genomes of its progenitors, with little recombination between homoeologs and only minor loss of genes (<5%). Most of the homoeologous genes in white clover have not undergone silencing, as similar proportions of genes are expressed in all three species. This is similar to some recent polyploids, but in contrast to others that have undergone major gene loss and silencing, indicating the need for more research on the factors driving gene loss in polyploids. Identification of polymorphisms specific to each progenitor allowed the authors to examine whether one homoeologous gene showed higher expression than its counterpart. About 69% of genes showed such bias, which was consistent across tissues. The genes that did not show consistent ratios across tissues might include loci that have undergone selection for expression patterns that provide a fitness advantage—examination of these genes showed that they were enriched in genes related to phenylpropanoid biosynthesis, particularly flavonoids. This fits nicely with the known roles of flavonoids in beneficial plant-microbial symbioses, and tolerance to biotic and abiotic stress; furthermore, it raises interesting questions for future work on gene expression in polyploids. Moreover, the observed differences in expression did not reflect differences in the progenitor species and therefore likely emerged after allopolyploidization.
In summary, this study provides new insight and opens major avenues for research on the longstanding question of whether and how polyploidy confers advantages on different species. Indeed, examination of how the genes within each subgenome respond to different environmental conditions, the resulting alterations in flavonoids and other metabolites, and what this means for adaptation by this species will give us a lot to think about as we look over our own patches of allotetraploid clover.
REFERENCES
Griffiths, A.G., Moraga, R., Tausen, M., Gupta, V., Bilton, T.P., Campbell, M.A., Ashby, R., Nagy, I., Khan, A., Larking, A., Anderson, C., Franzmayr, B., Hancock, K., Scott, A., Ellison, N.W., Cox, M.P., Asp, T., Mailund, T., Schierup, M.H., and Andersen, S.U. (2019). Breaking Free: the Genomics of Allopolyploidy-facilitated Niche Expansion in White Clover. Plant Cell DOI: https://doi.org/10.1105/tpc.18.00606.
Soltis, P.S., Marchant, D.B., Van de Peer, Y., and Soltis, D.E. (2015) Polyploidy and genome evolution in plants. Current Opinion in Genetics & Development, 35:119–125
Figure 1. Range of white clover and the extant relatives of its diploid progenitors
White clover (T. repens) occurs broadly in Europe and Asia (dark green) and has been introduced worldwide (light green). By contrast, the extant relatives of its diploid progenitors, T. occidentale (blue) and T. pallescens (orange), show a very restricted range. [Reprinted from Griffiths et al., 2019; Figure 1]




