Is Genetic Evolution Predictable?
Yang et al. investigate the evolution of flowering time in the young species Capsella rubella. https://doi.org/10.1105/tpc.18.00124
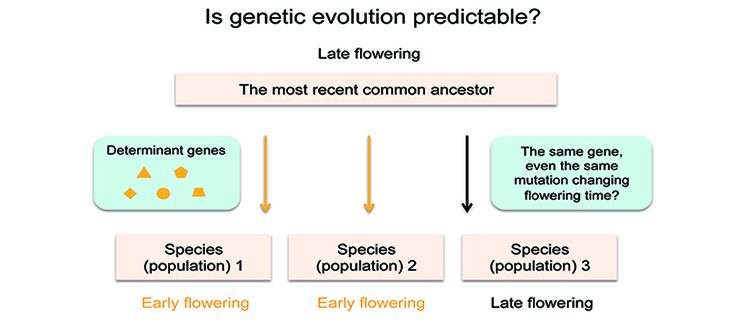
Background: Flowering time is an important adaptive life-history trait in plants. Capsella rubella, a close relative of Arabidopsis thaliana and—in evolutionary time—a young species, has reduced standing genetic variation due to an extreme bottleneck event (something that greatly reduced population size) early in its evolutionary history. Nonetheless, today, natural accessions (varieties within the species) of C. rubella display extensive variation for flowering time. Investigation of the molecular basis of flowering time variation in C. rubella therefore will broaden our understanding of the genetic factors regulating this variation in nature and also provide an excellent opportunity to understand how phenotypic diversity can occur with a limited initial gene pool.
Question: Our molecular understanding of how variation in flowering time is achieved is still rudimentary in species besides Arabidopsis. C. rubella is of particular interest because it went through an extreme genetic bottleneck during its speciation, which resulted in its current low genetic diversity. Thus it provides a unique opportunity to study parallel evolution in plants with limited standing genetic variation. The goal of our study is to explore how such wide variation in flowering time can occur in a species with limited genetic variation.
Findings: In this study, we demonstrate that common allelic variation and parallel evolution at the FLC locus (the same gene known to play a key role in the regulation of flowering time in Arabidopsis) confer variation in flowering time in C. rubella. We show that two overlapping deletions in the 5’ untranslated region of C. rubella FLC, which are associated with open chromatin state and histone modification levels, reduce its expression levels and promote flowering. More importantly, this study provides an example of how parallel mutations in the same 5’ untranslated region can shape phenotypic evolution in plants.
Next steps: It will be of great interest to clarify the extent to which parallel evolution shapes variation in other traits among natural populations of C. rubella, and more broadly, the role of parallel evolution in diverse taxa in general.
Li Yang, Hui-Na Wang, Xing-Hui Hou, Yu-Pan Zou, Ting-Shen Han, Xiao-Min Niu, Jie Zhang, Zhong Zhao, Marco Todesco, Sureshkumar Balasubramanian, and Ya-Long Guo. (2018). Parallel evolution of common allelic variants confers flowering diversity in Capsella rubella. Plant Cell https://doi.org/10.1105/tpc.18.00124.