Structure of KDM5 Gives Insight to Histone Demethylation
Yang et al. reveal a conserved H3K4me3 recognition mechanism shared by both plant and animal KDM5 histone demethylases. The Plant Cell (2017). https://doi.org/10.1105/tpc.17.00666
By Z. Yang, Q. Qiu, X. Cao and J. Du
Background: Histone methylation is a conserved gene regulation mechanism in plants and animals. The gene activation mark histone 3 lysine 4 trimethylation (H3K4me3) can be actively removed by specific enzymes – the KDM5 subfamily histone demethylases. In human, the KDM5 demethylases can promote tumor genesis and the development of drug tolerance of cancer cells. Therefore, human KDM5 enzymes are potent anti-cancer drug targets. In plants, one of the KDM5 subfamily enzyme, Arabidopsis thaliana JUMONJI 14 (AtJMJ14), functions in flowering regulation, RNA silencing and RNA-directed DNA methylation. However, the mechanism of the substrate specificity of the KDM5 enzymes, either from animals or plants, is still unknown.
Question: We wanted to know how the plant KDM5 subfamily H3K4me3 demethylase, especially AtJMJ14, specifically recognizes the substrate, and if the recognition mechanism is also conserved in animals.
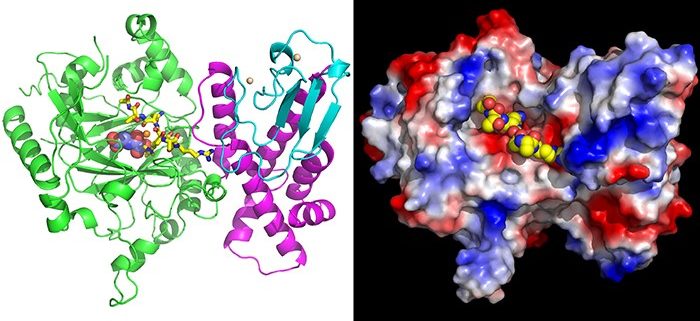
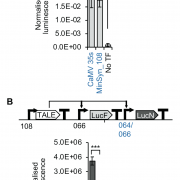
Findings: We determined the crystal structures of AtJMJ14 both in free form and in complex with the substrate H3K4me3 peptide. The structure analysis revealed that the H3R2 and H3Q5 are specifically recognized by several acidic residues of AtJMJ14, which provides the determinant of the substrate specificity of AtJMJ14. The mutations of these acidic residues significantly impair the activity of AtJMJ14 both in vitro and in vivo. By comparing the primary sequences and 3-dimentional structures of human and plant KDM5 proteins, we found that these critical acidic residues are not only conserved in plants, but also conserved in animal KDM5 subfamily demethylases. The in vitro demethylase assay towards human KDM5B showed that the mutations of the acidic residues can decrease the activity of human KDM5B, too, suggesting a conserved substrate recognition mechanism is shared by both plants and animals.
Next steps: The molecular mechanism of plant histone demethylase is largely unknown. Our work provides molecular insight into the substrate specificity of a plant KDM5 subfamily demethylase. In addition, human KDM5 demethylases are important anti-cancer drug targets, but the lack of KDM5-H3K4me3 complex structure restricts the development of KDM5-specific inhibitors. We demonstrated that the substrate recognition mechanism of AtJMJ14 is also conserved in human KDM5, thereby shedding light on structure-based KDM5-targeting inhibitor design.
Zhenlin Yang, Qi Qiu, Wei Chen, Bei Jia, Xiaomei Chen, Hongmiao Hu, Kaixuan He, Xian Deng, Sisi Li, W Andy Tao, XiaoFeng Cao, Jiamu Du (2017). Structure of Arabidopsis JMJ14-H3K4me3 complex reveal insight into the substrate specificity of KDM5 subfamily histone demethylase. Plant Cell. Published December 2017. DOI: https://doi.org/10.1105/tpc.17.0066