Getting to Know Each Cell in an Arabidopsis Root
Jean-Baptiste et al. use single-cell transcriptomic profiling to map developmental trajectories and assess cell-type specific changes during heat shock, demonstrating that single-cell transcriptomics holds promise for studying plant development and plant physiology at unprecedented resolution. Plant Cell https://doi.org/10.1105/tpc.18.00785
By Josh T. Cuperus and Ken Jean-Baptiste, Department of Genome Sciences, University of Washington, Seattle, WA, USA
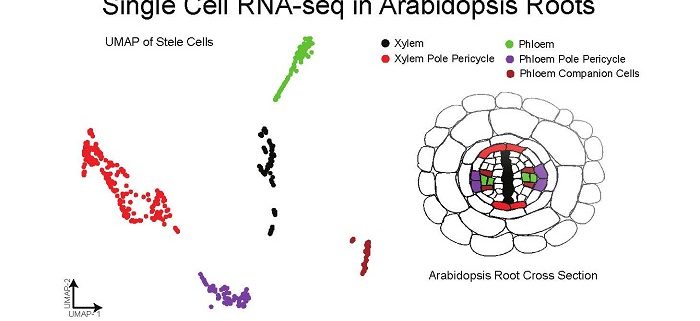
Background: Plants are made up of many different types of cells with different roles. These cells can be identified based on the types of genes they express, which are important to their roles. Many approaches have been used to study specific types of cells in Arabidopsis roots, including methods to express fluorescence proteins in only some types of cells. Newer technologies can capture thousands of individual cells at the same time, measuring the total RNA molecules in each cell. We have applied this single-cell RNA-seq technology to the Arabidopsis root.
Question: We sought to answer several important questions, but also bring new technological advances to plant systems. We asked the biological questions: Can distinct cell-types be resolved using single-cell RNA-seq of a whole Arabidopsis root? Can developmental information be understood using single-cell RNA-seq?
Findings: We identified all the major cell types in Arabidopsis roots based on each cell type’s unique RNA expression profile. We also visualized the developmental trajectory of many cell types, finding differences between young and old cells within the same tissue, and identifying transcription factors that were expressed specifically in young or old cells. We captured distinct, small populations of cells, including ones actively dividing. Lastly, we applied an abiotic stress, heat shock, to these roots. We found drastic changes in expression, some of which were cell-type specific. Cells in the exterior layers of the root had larger changes in expression than did interior cell types.
Next steps: Our work shows that it is possible to discern different cells with single cell RNA-seq in Arabidopsis roots, so in the future we hope to apply the same technique to Arabidopsis shoots, and other plants such as camelina, sorghum, and maize.
Ken Jean-Baptiste, José L. McFaline-Figueroa, Cristina M. Alexandre, Michael W. Dorrity, Lauren Saunders, Kerry L. Bubb, Cole Trapnell, Stanley Fields, Christine Queitsch, and Josh T. Cuperus. (2019). Dynamics of Gene Expression in Single Root Cells of A. thaliana. Plant Cell. https://doi.org/10.1105/tpc.18.00785.
Key words: single cell RNA-seq, Arabidopsis root cells, transcriptome, heat shock