Function of m6A in Arabidopsis
Wei et al. Study m6A binding protein ECT2 in Arabidopsis. The Plant Cell (2018). https://doi.org/10.1105/tpc.17.00934
By Lian-Huan Wei
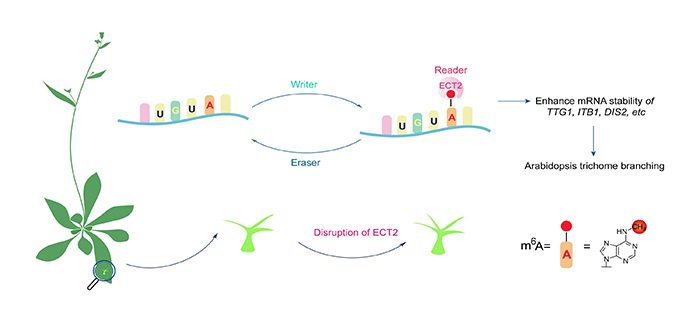
Background: N6-methyladenosine (m6A) is the most abundant chemical modification of eukaryotic messenger RNA (mRNA). The regulation of m6A modification plays important roles in plant development, but the mechanism is not completely understood. m6A is installed by the methyltransferase complex (“m6A writer”), removed by demethylases (“m6A erasers”), and recognized by certain binding proteins (“m6A readers”) to transduce its downstream regulatory effects, which include aspects of RNA processing. The m6A writer and eraser proteins in Arabidopsis have been discovered, but little is known about the m6A binding protein reader. Characterization of m6A binding proteins in Arabidopsis is essential for understanding m6A function and its role in plant growth and development.
Question: The Arabidopsis ECT2 protein contains a domain that is a well characterized m6A binding domain in mammals. Does ECT2 specifically recognize m6A in Arabidopsis – in other words, is ECT2 a plant m6A reader? If so, what RNA substrates and binding motif can be “read” by ECT2?
Findings: ECT2 is expressed in all tissues examined in Arabidopsis and strongly expressed in rapidly developing tissues. We show that ECT2 is an m6A binding protein and its m6A-binding function is required for trichome branching in Arabidopsis. Cellular localization studies showed that ECT2 is present in both the nucleus and the cytoplasm. Development of a formaldehyde cross-linking and immunoprecipitation method, called FA-CLIP, allowed us to identify more than 6000 ECT2 binding sites covering over 3,680 unique mRNA transcripts. We found that ECT2 preferentially binds the UGUA motif within the 3’ untranslated region (UTR) of mRNA transcripts, which is confirmed as a plant-specific m6A motif. Disruption of ECT2 accelerated the degradation of three ECT2-binding transcripts related to trichome morphogenesis, thereby affecting trichome branching.
Next steps: We will investigate the exact pathway underlying how mRNA stability is affected by ECT2’s m6A binding function. From the subcellular localization of ECT2 and sequencing data analysis, we speculate ECT2 could affect mRNA stability either by promoting m6A-mediated alternative polyadenylation of 3′ UTRs or by directly facilitating mRNA stability in the cytoplasm.
Lian-Huan Wei, Peizhe Song, Ye Wang, Zhike Lu, Qian Tang, Qiong Yu, Yu Xiao, Xiao Zhang, Hong-Chao Duan, Guifang Jia (2018). The m6A Reader ECT2 Controls Trichome Morphology by Affecting mRNA Stability in Arabidopsis Plant Cell May 2018, 30: 968-985; DOI: https://doi.org/10.1105/tpc.17.00934