Fear Not the Unknown: OPENER as a Study in Shedding Light on Genes with Unknown Function
Genome sequencing has become (relatively) cheap and easy, but assigning functions to the genes identified remains challenging—even in exhaustively studied species such as Arabidopsis thaliana, where functions of ~30% of genes remain unknown. Many of these genes likely have functions that are essential, and therefore, functional characterization is more challenging because loss of function of essential genes causes lethality in the seed or embryo (Muralla et al., 2011).
Examination of essential genes holds promise to identify previously unknown, essential metabolic, cellular, and developmental processes (Meinke et al., 2008)— how does one address this? Wang et al. (2019) start by focusing on conserved genes in Arabidopsis. Moreover, they narrow this focus to genes that are single copy and tend to remain single copy during evolution, despite the many whole-genome duplications that have occurred in various lineages. For these genes, the presence of multiple paralogs may have some disadvantage, such as dosage effects on gene expression, or the potential for dominant negative mutations that kill a protein complex (reviewed in Panchy et al, 2016). The authors found 5279 nuclear genes annotated as having unknown molecular function and involved in an unknown biological process. Of these, 194 single-copy genes were conserved in Arabidopsis, Physcomitrella patens, and Chlamydomonas reinhardtii.
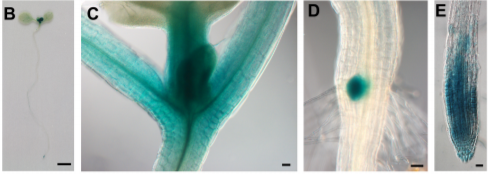
For essential genes, loss-of-function is expected to cause lethality at early stages of development (Muralla et al., 2011); therefore, the authors focused on one locus that is expressed in the root tip and for which T-DNA insertion mutants showed an early seed abortion phenotype. Because the arrested embryos looked like bottle openers, the authors named the locus OPENER (OPNR). Indeed, the opnr mutant embryos arrested very early in development and showed enlarged nucleoli and vacuoles. Moreover, transmission ratios indicated a potential defect in female gametophyte development.
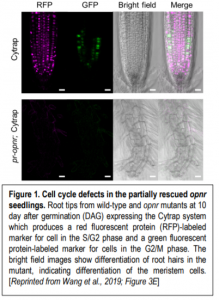 So, OPNR is essential—but for what? To answer this, the authors used RT-PCR and promoter-reporter fusion constructs to examine OPNR expression, finding that it is expressed in tissues where cells are actively dividing, such as the shoot meristem and root tip. To produce mutant seedlings, the authors partially rescued the mutant phenotype by expressing OPNR under a seed-specific promoter. Staining with the cell cycle marker Cell Cycle Tracking in Plant Cells (Cytrap) system showed that the partially rescued opnr root meristems had cell cycle defects and eventually differentiated rather than continuing to divide (figure).
So, OPNR is essential—but for what? To answer this, the authors used RT-PCR and promoter-reporter fusion constructs to examine OPNR expression, finding that it is expressed in tissues where cells are actively dividing, such as the shoot meristem and root tip. To produce mutant seedlings, the authors partially rescued the mutant phenotype by expressing OPNR under a seed-specific promoter. Staining with the cell cycle marker Cell Cycle Tracking in Plant Cells (Cytrap) system showed that the partially rescued opnr root meristems had cell cycle defects and eventually differentiated rather than continuing to divide (figure).
Immunoprecipitation and mass spectrometry-based assays identified OPNR interactors, including the nuclear envelope Sad1/UNC-84 (SUN) domain proteins SUN1 and SUN2. Indeed, OPNR fused to a fluorescent reporter localized to the nuclear envelope and this localization required SUN1 and SUN2. Staining with MitoTracker showed that OPNR also unexpectedly localized to the mitochondrial membrane and this was independent of SUN1 and SUN2. Consistent with this, OPNR co-immunoprecipitated with the mitochondrial proteins PROHIBITIN3 and PROHIBITIN4 and the partially rescued opnr mutants had smaller mitochondria than wild type. Other OPNR interactors have potential functions in ubiquitin-mediated protein degradation, indicating a possible role for OPNR in protein turnover.
Elucidation of OPNR function will require further research to piece together these intriguing observations. Nevertheless, this study provides a striking example of how researchers can bring to bear the formidable array of community resources, including libraries of insertional mutants, expression data, markers, and annotated genome sequence data across the plant kingdom, to provide insight into a novel process and previously uncharacterized protein. We can only anticipate the additional discoveries that await us in the unknown.
REFERENCES
Meinke, D., Muralla, R., Sweeney, C., and Dickerman, A. (2008). Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 13: 483-491
Muralla, R., Lloyd, J., and Meinke, D. (2011). Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS One 6: e28398.
Panchy, N., Lehti-Shiu, M., and Shiu, S.H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171: 2294-2316.
Wang, W., Zhang, X., and Niittylä, T. (2019). OPENER Is a Nuclear Envelope and Mitochondria Localized Protein Required for Cell Cycle Progression in Arabidopsis. Plant Cell DOI: https://doi.org/10.1105/tpc.19.00033.