Connecting the SUMO Ligase SIZ1 to Its Targets by Differential Proteomics
Rytz et al. investigate the selectivity of SUMOylation in Arabidopsis. Plant Cell (2018) https://doi.org/10.1105/tpc.17.00993
By Thérèse C. Rytz and Richard D. Vierstra
Background: The covalent attachment of small ubiquitin-like modifier (SUMO) to other intracellular proteins is an essential modification in plants that not only regulates growth and development, but also plays a central role in defense against numerous environmental challenges, such as extremes of heat and cold, drought, and pathogen attack. Upon experiencing heat stress, SUMO is rapidly conjugated to numerous proteins, most of which are localized to the nucleus and control a wide range of nuclear functions. SUMOylation occurs through a three-step reaction cascade with the final step directed by SUMO ligases that select specific substrates. In Arabidopsis thaliana, a small set of ligases is currently known, which include SAP-MIZ domain-containing 1 (SIZ1) and METHYL METHANESULFONATE-SENSITIVE21 (MMS21). While a few hundred SUMO targets have been identified in plants, it is unclear which ligases are responsible for modifying which proteins.
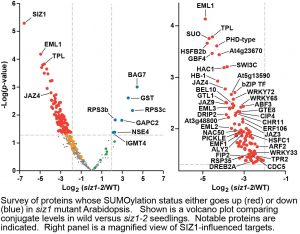
Questions: We sought to determine the substrates of the SUMO ligases SIZ1 and MMS21 using a large-scale mass spectrometric approach. To identify these targets, we identified over a thousand SUMOylated proteins in wild-type Arabidopsis and compared their levels to those in siz1 or mms21 mutants.
Findings: Whereas we were unable to definitively assign targets to MMS21, suggesting this ligase modifies only a small subset of low-abundance proteins, we could assign over 100 targets to SIZ1 upon heat stress. Most of these SIZ1-dependent substrates reside in the nucleus, and include well-known transcription factors, coactivators/repressors, and chromatin modifiers connected to abiotic and biotic stress defense. Additionally, SIZ1 selectively SUMOylates members of the TOPLESS corepressor family and multiple components of the SWI-SNF chromatin-remodeling complex, thus implicating both SUMO and SIZ1 as crucial regulators of chromatin structure/function and transcription. Collectively, SUMOylation by the SIZ1 ligase likely provides broad stress protection by modifying a large array of key nuclear regulators.
Next steps: Our work provides a deeper understanding of how SUMOylation protects plants against environmental stress. Further studies on these SUMO targets, including identification of SUMO attachment sites and understanding the impact of the SUMO moiety on target function, should reveal how SUMOylation provides stress protection, which can be used to enhance stress tolerance for agricultural benefit.
Thérèse C. Rytz, Marcus J. Miller, Fionn McLoughlin, Robert C. Augustine, Richard S. Marshall, Yu-ting Juan, Yee-yung Charng, Mark Scalf, Lloyd M. Smith, Richard D. Vierstra (2018). SUMOylome Profiling Reveals a Diverse Array of Nuclear Targets Modified by the SUMO Ligase SIZ1 during Heat Stress. Plant Cell May 2018, 30: 1077-1099; DOI: https://doi.org/10.1105/tpc.17.00993