Basal vs. Non-basal Polarity: Different Endomembrane Trafficking Pathways Establish Different Patterns
IN BRIEF: Nancy R. Hofmann nhofmann@aspb.org
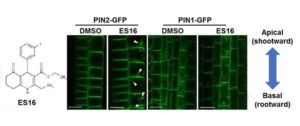 Plant development and responses to the environment hinge on the ability to target proteins to different areas of the plasma membrane within a cell. Indeed, the establishment of polar distributions of proteins such as PIN auxin transporters is among the key responsibilities of the endomembrane trafficking system (reviewed in Morita and Shimada, 2014). Although some features of trafficking are shared between proteins targeted to different portions of the plasma membrane, others are specific. For instance, GNOM, an ADP ribosylation factor guanine nucleotide exchange factor (ARF-GEF), appears to function in basal but not apical localization of PIN proteins (Kleine-Vehn et al., 2008). In new work, Li et al. (2017) develop and use endosidin 16 (ES16) as a pathway-specific tool to dissect the mechanisms underlying different patterns of polarity.
Plant development and responses to the environment hinge on the ability to target proteins to different areas of the plasma membrane within a cell. Indeed, the establishment of polar distributions of proteins such as PIN auxin transporters is among the key responsibilities of the endomembrane trafficking system (reviewed in Morita and Shimada, 2014). Although some features of trafficking are shared between proteins targeted to different portions of the plasma membrane, others are specific. For instance, GNOM, an ADP ribosylation factor guanine nucleotide exchange factor (ARF-GEF), appears to function in basal but not apical localization of PIN proteins (Kleine-Vehn et al., 2008). In new work, Li et al. (2017) develop and use endosidin 16 (ES16) as a pathway-specific tool to dissect the mechanisms underlying different patterns of polarity.
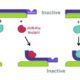
From a library of chemicals that disrupt endomembrane trafficking (Drakakaki et al., 2011), Li et al. identified ES16 as a compound that alters apical (shootward) localization of PIN2 in Arabidopsis thaliana root epidermis cells without affecting basal (rootward) localization of PIN1 (see Figure). ES16 treatment induced endomembrane bodies containing PIN2, indicative of disruption of trafficking, but did not alter the basal targeting of PIN1 in the root epidermis or of PIN3 or PIN7 in the stele.
While ES16 did not disrupt basal polarity, it did alter both lateral and non-polar localizations in addition to apical polarity. When the authors checked endocytosis and recycling of plasma membrane proteins, they found no effect of ES16 on uptake of FM4-64 dye (i.e., endocytosis) or on basal recycling of PIN1. By contrast, ES16 blocked recycling of PIN2 (apical polarity), PIP2a (non-polar), and NIP5;1 (lateral polarity). Thus, rather than defining a pathway specific for apical polarity, ES16 appears to target a general pathway used for non-basal targeting.
In agreement with its effects on the localization of PIN2, the phenotypes associated with ES16 treatment included inhibition of root elongation and root gravitropism. However, ES16 did not alter lateral root initiation, which depends on basal recycling of P1N1, consistent with a selective effect of ES16 on targeting pathways other than basal.
Further analysis indicated that ES16 likely disrupts a pathway that involves trafficking from the trans-Golgi network to the pre-vacuolar compartment and utilizes the RabA2A GTPase in parallel with BIG clade ARF-GEFs to modulate non-basal trafficking. In fact, a dominant negative mutation of RabA2A had similarly specific effects on PIN2 (and not PIN1) localization as ES16 treatment.
This work points to a biologically relevant distinction between basal and non-basal polarity targeting and provides an excellent tool for further dissection of post-Golgi trafficking to different areas of the plasma membrane.
REFERENCES
Drakakaki, G., Robert, S., Szatmari, A.M., Brown, M.Q., Nagawa, S., Van Damme, D., Leonard, M., Yang, Z., Girke, T., Schmid, S.L., Russinova, E., Friml, J., Raikhel, N.V., and Hicks, G.R. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sciences USA 108: 17850-17855.
Kleine-Vehn, J., Dhonukshe, P., Sauer, M., Brewer, P.B., Wisniewska, J., Paciorek, T., Benkova, E., and Friml, J. (2008b). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Current biology : CB 18: 526-531.
Morita, M.T., and Shimada, T. (2014). The plant endomembrane system–a complex network supporting plant development and physiology. Plant Cell Physiol. 55: 667-671.
Li, R., Rodriguez-Furlan, C., Wang, J., van de Ven, W., Gao, T., Raikhel, N.V., and Hicks, G.R. (2017). Different Endomembrane Trafficking Pathways Establish Apical and Basal Polarities. Plant Cell. doi: 10.1105/tpc.16.00524.


Leave a Reply
Want to join the discussion?Feel free to contribute!