A Maize Mutant Modifier Links Development and Defense
Anderson et al. identify a second site modifier that rescues a maize developmental mutant and report that it encodes a homolog of Arabidopsis ENHANCED DISEASE RESISTANCE4, providing insight into the integration of developmental control and immune responses. Plant Cell https://doi.org/10.1105/tpc.18.00840
By Alyssa Anderson, Sarah Hake
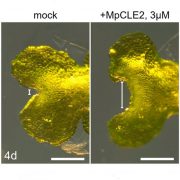
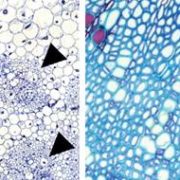
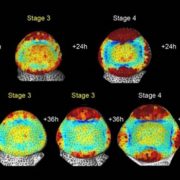
Background: The progression from a seed to a full-grown plant is an intricate process that demands coordination of many networks, such as hormonal, metabolic, and defense pathways. Maize is a particularly rich system for exploring developmental programming in part due to the high levels of genetic diversity found within the species. Liguleless narrow (Lgn-R) is a mutant that inhibits proper formation of diverse tissues, including the leaves, stalk, ears and tassel. However, this phenotype is only present in some genetic backgrounds of maize and the same mutation leads to few noticeable phenotypes in other backgrounds. The mutation eliminates the kinase activity of the Lgn-R protein, presumably altering the downstream signaling cascades of its targets and somehow resulting in the observed developmental phenotypes.
Question: In a previous study we were able to narrow down the location of one of the background dependent modifiers of Lgn-R to a region on Chromosome 1. Here we set out to discover exactly which gene was responsible for these background differences and determine what pathways were involved in the severe Lgn-R developmental phenotype.
Findings: By analyzing sequence and expression data and conducting a mutagenesis screen, we were able to pinpoint our Lgn-R modifier, Sympathy for the ligule (Sol), as a maize homolog of Arabidopsis ENHANCED DISEASE RESISTANCE4 (EDR4). Sol has increased expression exclusively in the severe mutant backgrounds and, like EDR4, has increased expression in response to treatment with pathogen associated molecular patterns (PAMPs). We also analyzed large-scale expression and phosphorylation differences in the Lgn-R background and found additional evidence of mis-regulation of immune pathways in the mutant, which may or may not be distinct from the impacted developmental pathways. Sol is capable of interacting with these pathways in a way that leads to extreme phenotypic differences across the diversity of our maize backgrounds.
Next steps: Current work with collaborators aims to determine if Lgn-R also has a disease resistance phenotype when under pathogen attack. We also hope to elucidate the exact molecular mechanism of Sol and how it mitigates our severe developmental phenotypes.
Alyssa Anderson, Brian St. Aubin, María Jazmín Abraham-Juárez, Samuel Leiboff, Zhouxin Shen, Steve Briggs, Jacob O. Brunkard, Sarah Hake. (2019). The second site modifier, Sympathy for the ligule, encodes a homolog of Arabidopsis ENHANCED DISEASE RESISTANCE4 and rescues the Liguleless narrow maize mutant. Plant Cell. https://doi.org/10.1105/tpc.18.00840.
Key words: maize, development, immunity, ligule