What We’re Reading: September 22nd
We had a large number of guest submissions to What We’re Reading this week, thank you to all of the contributors! We even had two submissions of the same paper, so we’ve included both – clearly it’s a good paper to have a look at!
Review: Multiple routes of light signaling during root photomorphogenesis
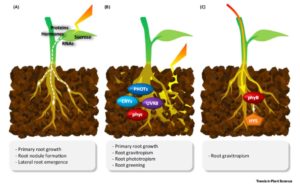 Light response research in plants has traditionally focused on the shoot, but recent studies have revealed that roots are also light responsive. Lee et al. address the why and how of root photomorphogenesis. They review three ways that light is perceived in roots: via mobile signals from the shoot, direct sensing of light by root cells, and light channeling through the plant body. Interestingly, stem-piped light is highly enriched for the near infra-red (IR) and IR parts of the spectrum. As yet nothing is known about IR light perception in plants. However, the molecular basis for IR light perception in snakes is known, and the authors show that similar genes as those involved in snake IR perception are present in plants, although these genes have not yet been functionally defined in plants. Who knows, maybe roots and rattlesnakes have more in common than their shapes? Trends Plant Sci. 10.1016/j.tplants.2017.06.009
Light response research in plants has traditionally focused on the shoot, but recent studies have revealed that roots are also light responsive. Lee et al. address the why and how of root photomorphogenesis. They review three ways that light is perceived in roots: via mobile signals from the shoot, direct sensing of light by root cells, and light channeling through the plant body. Interestingly, stem-piped light is highly enriched for the near infra-red (IR) and IR parts of the spectrum. As yet nothing is known about IR light perception in plants. However, the molecular basis for IR light perception in snakes is known, and the authors show that similar genes as those involved in snake IR perception are present in plants, although these genes have not yet been functionally defined in plants. Who knows, maybe roots and rattlesnakes have more in common than their shapes? Trends Plant Sci. 10.1016/j.tplants.2017.06.009
Review: Transcriptional control of photosynthetic capacity – conservation and divergence from Arabidopsis to rice
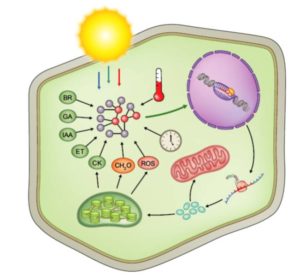 About 3000 genes are required for a plant to carry out photosynthesis. Wang et al. review the transcriptional control of these photosynthetic genes, drawing on transcriptomic and evolutionary studies to make comparisons between Arabidopsis and grasses. Photosynthesis of course starts with light, and the authors’ review starts with photoreceptors and their downstream transcription factors, focusing on lessons from Arabidopsis. Other sections focus on the transcriptional regulators involved in chloroplast development, communication between chloroplast and nucleus, and environmental responses. The second part of the review looks at similarities and differences in these gene regulatory networks in rice. As the authors point out, extending beyond Arabidopsis is crucial to address future food demands using systems and synthetic biology applications in crop plants. New Phytol. 10.1111/nph.14682
About 3000 genes are required for a plant to carry out photosynthesis. Wang et al. review the transcriptional control of these photosynthetic genes, drawing on transcriptomic and evolutionary studies to make comparisons between Arabidopsis and grasses. Photosynthesis of course starts with light, and the authors’ review starts with photoreceptors and their downstream transcription factors, focusing on lessons from Arabidopsis. Other sections focus on the transcriptional regulators involved in chloroplast development, communication between chloroplast and nucleus, and environmental responses. The second part of the review looks at similarities and differences in these gene regulatory networks in rice. As the authors point out, extending beyond Arabidopsis is crucial to address future food demands using systems and synthetic biology applications in crop plants. New Phytol. 10.1111/nph.14682
Review: Naphthylphthalamic acid and the mechanism of polar auxin transport ($)
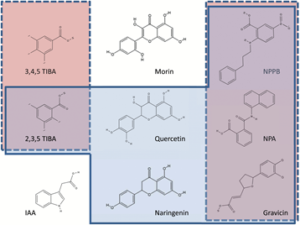 Teale and Palme give an overview of what the last 60 years of using synthetic chemicals inhibitors of polar auxin transport has really taught us. Key among inhibitors is Naphthylphthalamic acid (NPA) that interferes with polar auxin transport and can mimic PIN gene mutant phenotypes. These inhibitors have proven to be invaluable tools and have made it possible to uncover as much as we know today about how auxin is moved, through their complex interactions with the different auxin transporters. Yet questions about auxin transport still remain, including specifics of inhibitor binding, roles of phosphorylation, and whether endogenous plant inhibitors function equivalently. The authors argue that perhaps the clues these inhibitors have and still can provide will lead us to the solutions. (Summary by Aaron Rashotte) J. Exp. Bot. 10.1093/jxb/erx323
Teale and Palme give an overview of what the last 60 years of using synthetic chemicals inhibitors of polar auxin transport has really taught us. Key among inhibitors is Naphthylphthalamic acid (NPA) that interferes with polar auxin transport and can mimic PIN gene mutant phenotypes. These inhibitors have proven to be invaluable tools and have made it possible to uncover as much as we know today about how auxin is moved, through their complex interactions with the different auxin transporters. Yet questions about auxin transport still remain, including specifics of inhibitor binding, roles of phosphorylation, and whether endogenous plant inhibitors function equivalently. The authors argue that perhaps the clues these inhibitors have and still can provide will lead us to the solutions. (Summary by Aaron Rashotte) J. Exp. Bot. 10.1093/jxb/erx323
Review: Sensing danger – key to activating plant immunity
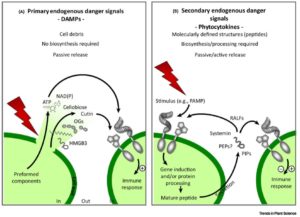 The first step in defending yourself is recognizing that you need to defend yourself. Gust et al. review the mechanisms through which plants sense danger, drawing parallels to similar mechanisms in animals. They define three categories of danger signals. Exogenous signals are “non-self” signals, such as the pathogen-associated molecular patterns (PAMPS) or signals produced by herbivores, nematodes, or parasitic plants, as well as their effectors. Endogenous or self signals fall into two classes, the primary damage-associated molecular patterns (DAMPS, e.g., cell wall fragments), and the secondary signals produced in response to danger (e.g., processed peptides, also described as phytocytokines by analogy to animal cytokines). A third class, abiotic danger signals (e.g., nanomaterials), is less well defined in plants. This interesting and accessible review should help students appreciate the complexity of plant defense mechanisms. Trends Plant Sci. 10.1016/j.tplants.2017.07.005
The first step in defending yourself is recognizing that you need to defend yourself. Gust et al. review the mechanisms through which plants sense danger, drawing parallels to similar mechanisms in animals. They define three categories of danger signals. Exogenous signals are “non-self” signals, such as the pathogen-associated molecular patterns (PAMPS) or signals produced by herbivores, nematodes, or parasitic plants, as well as their effectors. Endogenous or self signals fall into two classes, the primary damage-associated molecular patterns (DAMPS, e.g., cell wall fragments), and the secondary signals produced in response to danger (e.g., processed peptides, also described as phytocytokines by analogy to animal cytokines). A third class, abiotic danger signals (e.g., nanomaterials), is less well defined in plants. This interesting and accessible review should help students appreciate the complexity of plant defense mechanisms. Trends Plant Sci. 10.1016/j.tplants.2017.07.005
Engineering quantitative trait variation for crop improvement by genome editing ($)
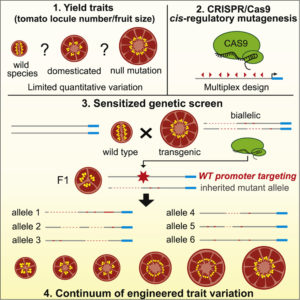 Variation is the engine of evolution, and plant breeders and geneticists have long relied on induced variation to create a population from which to select. Rodríguez-Leal et al. used CRISPR/Cas9 mutagenesis to selectively introduce variability into the promoter regions of three genes involved in tomato fruit size [including locule number (seed compartments)] and growth habit. Their system allows the transgenes to be segregated away, leaving non-transgenic, stable lines. Their strategy resulted in a set of alleles leading to a continuum of locule numbers and fruit size. This study is not only a proof-of-concept of a strategy to introduce targeted variation for breeding purposed, but also revealed the unexpected result that the expression level of key regulatory genes does not strictly correlate with phenotypic effects, supporting recent models for non-linear responses to dose-sensitive genes. Cell 10.1016/j.cell.2017.08.030
Variation is the engine of evolution, and plant breeders and geneticists have long relied on induced variation to create a population from which to select. Rodríguez-Leal et al. used CRISPR/Cas9 mutagenesis to selectively introduce variability into the promoter regions of three genes involved in tomato fruit size [including locule number (seed compartments)] and growth habit. Their system allows the transgenes to be segregated away, leaving non-transgenic, stable lines. Their strategy resulted in a set of alleles leading to a continuum of locule numbers and fruit size. This study is not only a proof-of-concept of a strategy to introduce targeted variation for breeding purposed, but also revealed the unexpected result that the expression level of key regulatory genes does not strictly correlate with phenotypic effects, supporting recent models for non-linear responses to dose-sensitive genes. Cell 10.1016/j.cell.2017.08.030
Regulation of rice root development by a retrotransposon acting as a microRNA sponge
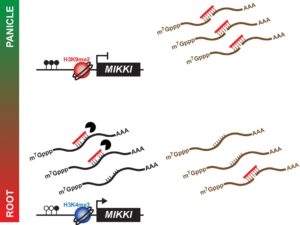 Transposable elements (TEs) compose a considerable portion of most plant genomes, and mounting evidence shows various roles of TEs in the regulation of gene expression. Some TE transcripts have been hypothesized to work as “sponges” to help fine-tune the levels of miRNAs through complementary binding. Though the miRNA sponge model is supported by TE overexpression studies, specific examples of its involvement in regulatory networks have yet to be seen. Here, Cho and Paszkowski report the novel role of a retrotransposon as a miRNA sponge in root development. They demonstrate that a root-specific transcript derived from a retrotransposon, dubbed MIKKI, acts as a decoy target for the miRNA miR171. miR171 is well-studied for its role in targeting members of the SCARECROW-LIKE family of root-specific transcription factors. MIKKI transcripts demonstrate modulation of miR171 levels in roots, thus revealing an additional tier of complexity to this transcriptional network and a novel mechanism of gene regulation. (Summary by Alexander Meyers) eLIFE 10.7554/eLife.30038
Transposable elements (TEs) compose a considerable portion of most plant genomes, and mounting evidence shows various roles of TEs in the regulation of gene expression. Some TE transcripts have been hypothesized to work as “sponges” to help fine-tune the levels of miRNAs through complementary binding. Though the miRNA sponge model is supported by TE overexpression studies, specific examples of its involvement in regulatory networks have yet to be seen. Here, Cho and Paszkowski report the novel role of a retrotransposon as a miRNA sponge in root development. They demonstrate that a root-specific transcript derived from a retrotransposon, dubbed MIKKI, acts as a decoy target for the miRNA miR171. miR171 is well-studied for its role in targeting members of the SCARECROW-LIKE family of root-specific transcription factors. MIKKI transcripts demonstrate modulation of miR171 levels in roots, thus revealing an additional tier of complexity to this transcriptional network and a novel mechanism of gene regulation. (Summary by Alexander Meyers) eLIFE 10.7554/eLife.30038
The G-box transcriptional regulatory code in Arabidopsis
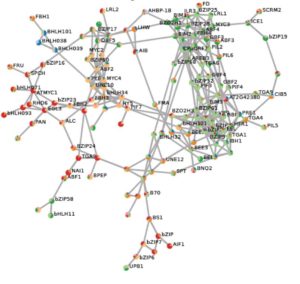 The G-box (CACGTG) is a DNA element widespread in plant genomes and recognized by two large families of transcription factors (TFs), the basic leucine zipper family (bZIP) and the basic-helix-loop-helix (bHLH) family. Members of these TF families contribute to growth, temperature and light signaling, and drought and immune responses. Collectively there are more than 170 genes encoding members of these two TF families in Arabidopsis. The proteins can bind to their target sequences as homo- or hetero-dimers, which makes matching TFs with their target genes quite complicated. Ezer et al. used in vitro binding and DAP-seq data as well as a large set of RNA-seq time course data to construct a gene regulatory network that identifies the set of bZIPs and bHLHs that are most predictive of expression of genes downstream of perfect G-boxes, and used the network to correctly predict the effects of knocking-out a PIF transcription factor. The Ara-BOX-cis network is available for use by the community. Plant Physiol. 10.1104/pp.17.01086
The G-box (CACGTG) is a DNA element widespread in plant genomes and recognized by two large families of transcription factors (TFs), the basic leucine zipper family (bZIP) and the basic-helix-loop-helix (bHLH) family. Members of these TF families contribute to growth, temperature and light signaling, and drought and immune responses. Collectively there are more than 170 genes encoding members of these two TF families in Arabidopsis. The proteins can bind to their target sequences as homo- or hetero-dimers, which makes matching TFs with their target genes quite complicated. Ezer et al. used in vitro binding and DAP-seq data as well as a large set of RNA-seq time course data to construct a gene regulatory network that identifies the set of bZIPs and bHLHs that are most predictive of expression of genes downstream of perfect G-boxes, and used the network to correctly predict the effects of knocking-out a PIF transcription factor. The Ara-BOX-cis network is available for use by the community. Plant Physiol. 10.1104/pp.17.01086
Completing the whole puzzle of whole genome duplications in land plants
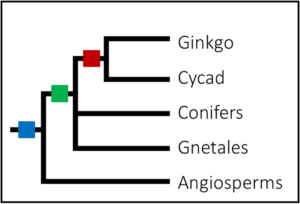 A hot topic in plant evolutionary biology is whole genome duplications (WGDs), in which an organism copies its entire genetic dataset. Having double the required DNA is often viewed as detrimental but can be useful in times of rapid environmental change. Recently, the role of WGDs during plant evolution was reviewed, further linking times of environmental uncertainty, such as the K-Pg extinction that led to the demise of the dinosaurs, to the apparently synchronous diversification in the flowering angiosperms. In the other, often neglected, lineages of land plants, such as the non-flowering gymnosperms of which only around ~1 000 extant species remain, evidence for WDGs are less prevalent. In 2015, a study largely focusing on the conifers identified duplication events that may have coincided with the Permian-Triassic boundary, the most severe mass extinction event in Earth’s history, and to which the survival of the conifers may be accredited. Additionally, the authors found a relatively recent WGD in one genera of the Gnetales lineage, Welwitschia, but no such event in the other extant genera of this lineage was identified. Evidence for WGDs in the remaining two gymnosperm lineages, Ginkgo biloba (the sole survivor of the Ginkgoales lineage) and the cycads remained uncertain. Recently however, the search for WGDs in all lineages of land plants identified an ancient WGD in G. biloba and the cycads. Interestingly, this duplication event seemed to be shared between these two lineages, as it appeared to have occurred in an ancestor of both the cycads and ginkgo before speciation occurred. What is even more remarkable is that the remnants of such ancient events can still be identified in the genomes of species alive today, indicative of both the slow evolutionary rate of some organisms and the value of high quality genetic data.(Summary by Danielle Roodt Prinsloo) PLOS ONE. 10.1371/journal.pone.0184454
A hot topic in plant evolutionary biology is whole genome duplications (WGDs), in which an organism copies its entire genetic dataset. Having double the required DNA is often viewed as detrimental but can be useful in times of rapid environmental change. Recently, the role of WGDs during plant evolution was reviewed, further linking times of environmental uncertainty, such as the K-Pg extinction that led to the demise of the dinosaurs, to the apparently synchronous diversification in the flowering angiosperms. In the other, often neglected, lineages of land plants, such as the non-flowering gymnosperms of which only around ~1 000 extant species remain, evidence for WDGs are less prevalent. In 2015, a study largely focusing on the conifers identified duplication events that may have coincided with the Permian-Triassic boundary, the most severe mass extinction event in Earth’s history, and to which the survival of the conifers may be accredited. Additionally, the authors found a relatively recent WGD in one genera of the Gnetales lineage, Welwitschia, but no such event in the other extant genera of this lineage was identified. Evidence for WGDs in the remaining two gymnosperm lineages, Ginkgo biloba (the sole survivor of the Ginkgoales lineage) and the cycads remained uncertain. Recently however, the search for WGDs in all lineages of land plants identified an ancient WGD in G. biloba and the cycads. Interestingly, this duplication event seemed to be shared between these two lineages, as it appeared to have occurred in an ancestor of both the cycads and ginkgo before speciation occurred. What is even more remarkable is that the remnants of such ancient events can still be identified in the genomes of species alive today, indicative of both the slow evolutionary rate of some organisms and the value of high quality genetic data.(Summary by Danielle Roodt Prinsloo) PLOS ONE. 10.1371/journal.pone.0184454
Epigenetic memory restoration and maintenance after cell division
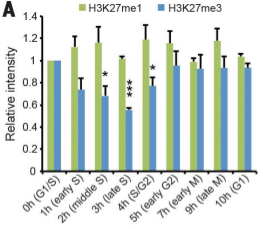 Nucleosomes, composed of histones, pack chromatin and make genes less available for transcription. Methylation of some histone positions is associated with repression of transcription and epigenetic silencing of some genes. After cell division during the cell cycle, maintenance of expression patterns requires restoration of those epigenetic marks after DNA replication in the G2 phase. Jiang and Berger show that trimethylation of H3.1 is restored early in G2 and remains stable in G1, and that this rapid restoration is specific to plants. Furthermore, proteins of the Polycomb complex are involved in the deposition and maintenance of the trimethylation marks, so have an important role on epigenetic silencing memory in response to the environment and potentially allowing for developmental plasticity. (Summary by Gaby Auge) Science 10.1126/science.aan4965
Nucleosomes, composed of histones, pack chromatin and make genes less available for transcription. Methylation of some histone positions is associated with repression of transcription and epigenetic silencing of some genes. After cell division during the cell cycle, maintenance of expression patterns requires restoration of those epigenetic marks after DNA replication in the G2 phase. Jiang and Berger show that trimethylation of H3.1 is restored early in G2 and remains stable in G1, and that this rapid restoration is specific to plants. Furthermore, proteins of the Polycomb complex are involved in the deposition and maintenance of the trimethylation marks, so have an important role on epigenetic silencing memory in response to the environment and potentially allowing for developmental plasticity. (Summary by Gaby Auge) Science 10.1126/science.aan4965
Stages of silencing to hold epigenetic memory
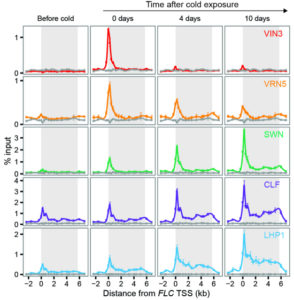 The gene FLC (FLOWERING LOCUS C) is a repressor of flowering in Arabidopsis thaliana. It needs to be silenced after exposure to prolonged cold for plants to be able to flower. The memory of winter leads to changes in the methylation state of Histone 3 associated with FLC in two steps: first by recruiting H3K27m3 (trimethylated H3 on lysine 27) in a nucleation region close to the transcriptional start site of FLC, and then by spreading of H3K27m3 over the entire FLC locus when plants are returned to warm conditions (after experiencing winter). Yang et al. show how key molecular players of the Polycomb Repressive Complex 2—namely VRN2, VIN3, VRN5, CLF, SWN and LHP1—are sequentially required at different stages of FLC silencing. VIN3, VRN2, VRN5 and SWN are required for nucleation of H3K27m3 during cold exposure at the transcriptional start site (also showing reversibility), while LHP1 and CLF are involved in H3K27m3 spreading after cold for long-term stable silencing. DNA replication during cell division impairs the stability of nucleation and is required for spreading of the epigenetic mark along the locus. Stability of the epigenetic state could be observed in cell files in the root, where daughter cells inherit the memory of FLC silencing in cis. Altogether, these results show a dynamic mechanism that could allow for highly plastic responses to environmental changes. (Summary by Gaby Auge) Science 10.1126/science.aan1121
The gene FLC (FLOWERING LOCUS C) is a repressor of flowering in Arabidopsis thaliana. It needs to be silenced after exposure to prolonged cold for plants to be able to flower. The memory of winter leads to changes in the methylation state of Histone 3 associated with FLC in two steps: first by recruiting H3K27m3 (trimethylated H3 on lysine 27) in a nucleation region close to the transcriptional start site of FLC, and then by spreading of H3K27m3 over the entire FLC locus when plants are returned to warm conditions (after experiencing winter). Yang et al. show how key molecular players of the Polycomb Repressive Complex 2—namely VRN2, VIN3, VRN5, CLF, SWN and LHP1—are sequentially required at different stages of FLC silencing. VIN3, VRN2, VRN5 and SWN are required for nucleation of H3K27m3 during cold exposure at the transcriptional start site (also showing reversibility), while LHP1 and CLF are involved in H3K27m3 spreading after cold for long-term stable silencing. DNA replication during cell division impairs the stability of nucleation and is required for spreading of the epigenetic mark along the locus. Stability of the epigenetic state could be observed in cell files in the root, where daughter cells inherit the memory of FLC silencing in cis. Altogether, these results show a dynamic mechanism that could allow for highly plastic responses to environmental changes. (Summary by Gaby Auge) Science 10.1126/science.aan1121
Natural variation of photosynthetic traits for enhanced yield in rice
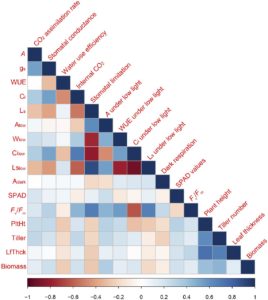 Natural variation in photosynthetic traits can be exploited for new targets for breeding or genetic engineering of crops. In an effort to identify traits which can lead to enhanced biomass production and therefore yield potential in rice, Qu et al. conducted a comprehensive survey of 14 photosynthetic parameters and four morphological traits of a global rice population under high- and low light conditions. The results revealed that the photosynthetic rate under low light highly correlates with biomass accumulation in rice. Notably, this important physiological trait is highly heritable, thus ready for genetic manipulation in contemporary rice breeding programs for better biomass production and yield potential. (Summary by Quang Vong Le) Plant Physiol. 10.1104/pp.17.00332
Natural variation in photosynthetic traits can be exploited for new targets for breeding or genetic engineering of crops. In an effort to identify traits which can lead to enhanced biomass production and therefore yield potential in rice, Qu et al. conducted a comprehensive survey of 14 photosynthetic parameters and four morphological traits of a global rice population under high- and low light conditions. The results revealed that the photosynthetic rate under low light highly correlates with biomass accumulation in rice. Notably, this important physiological trait is highly heritable, thus ready for genetic manipulation in contemporary rice breeding programs for better biomass production and yield potential. (Summary by Quang Vong Le) Plant Physiol. 10.1104/pp.17.00332
Elevated CO2 increases N2 fixation and contributes to various yield responses of soybean
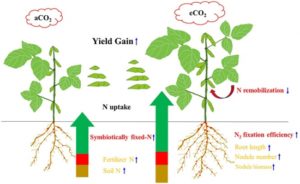 Climate change is certain to affect our ability to produce food crops and feed a growing global population in the twenty-first century. Free-air CO2 enrichment (FACE) experiments have shown that elevated CO2 (eCO2) levels, a major driver of climate change, have a consistently positive effect on the yield of the world’s most important crops, such as rice, wheat and soybean. However, under such conditions, nitrogen deficiency can become a limiting factor for plant productivity. Here, Li et al. explored nitrogen uptake mechanisms in eight soybean cultivars under eCO2. These cultivars varied in their response to eCO2, with seed yield increases ranging from 12 – 91%. Increased yield correlated with enhanced nitrogen uptake, primarily through symbiotic fixation, as well as a reduction in nitrogen remobilization from shoots to seeds during reproduction. This reduction in remobilization did not limit the seed nitrogen content. Specific interactions with the soil microbiome are likely to explain the observed differences between cultivars in increased nitrogen fixation and crop yield under eCO2. Research such as this will be essential to help achieve food security in a changing climate. (Summary by Mike T. Page) Front. Plant Sci. 10.3389/fpls.2017.01546.
Climate change is certain to affect our ability to produce food crops and feed a growing global population in the twenty-first century. Free-air CO2 enrichment (FACE) experiments have shown that elevated CO2 (eCO2) levels, a major driver of climate change, have a consistently positive effect on the yield of the world’s most important crops, such as rice, wheat and soybean. However, under such conditions, nitrogen deficiency can become a limiting factor for plant productivity. Here, Li et al. explored nitrogen uptake mechanisms in eight soybean cultivars under eCO2. These cultivars varied in their response to eCO2, with seed yield increases ranging from 12 – 91%. Increased yield correlated with enhanced nitrogen uptake, primarily through symbiotic fixation, as well as a reduction in nitrogen remobilization from shoots to seeds during reproduction. This reduction in remobilization did not limit the seed nitrogen content. Specific interactions with the soil microbiome are likely to explain the observed differences between cultivars in increased nitrogen fixation and crop yield under eCO2. Research such as this will be essential to help achieve food security in a changing climate. (Summary by Mike T. Page) Front. Plant Sci. 10.3389/fpls.2017.01546.
ABA-induced stomatal closure involves ALMT4, a phosphorylation-dependent vacuolar anion channel
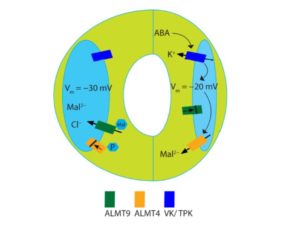 Changes in guard cell turgor pressure result in stomatal opening and closing, balancing CO2 uptake with transpiration. These dynamics have been studied as early as the 1800s and much knowledge has been gained regarding the components involved in this process, yet we are still far from a unified model and many questions still remain. Eisenach et al. identified an ion channel involved in the ABA-induced stomatal closure response pathway. ALMT4 is upregulated in guard cells with exposure to ABA and almt4 mutants are hyposensitive to ABA, displaying impairments in ABA-induced closure and ABA-inhibited opening. Electrophysiological characteristics analyzing the I-V (current-voltage) relationship of ALMT4 compared to introduced phophomimetic and dephosphomimetic mutants ALMT4 S382E and ALMT4 S382A, respectively, found that: ALMT4 serine-382 phosphorylation deactivates current activity; the dephosphomimetic mutation leads to constitutive activation; and ALMT4 is involved in the release of Mal2- from the vacuole to the cytoplasm of guard cells. These findings add another piece to the puzzle in our quest to understand stomatal dynamics. (Summary by Alecia Biel) Plant Cell 10.1105/tpc.17.00452
Changes in guard cell turgor pressure result in stomatal opening and closing, balancing CO2 uptake with transpiration. These dynamics have been studied as early as the 1800s and much knowledge has been gained regarding the components involved in this process, yet we are still far from a unified model and many questions still remain. Eisenach et al. identified an ion channel involved in the ABA-induced stomatal closure response pathway. ALMT4 is upregulated in guard cells with exposure to ABA and almt4 mutants are hyposensitive to ABA, displaying impairments in ABA-induced closure and ABA-inhibited opening. Electrophysiological characteristics analyzing the I-V (current-voltage) relationship of ALMT4 compared to introduced phophomimetic and dephosphomimetic mutants ALMT4 S382E and ALMT4 S382A, respectively, found that: ALMT4 serine-382 phosphorylation deactivates current activity; the dephosphomimetic mutation leads to constitutive activation; and ALMT4 is involved in the release of Mal2- from the vacuole to the cytoplasm of guard cells. These findings add another piece to the puzzle in our quest to understand stomatal dynamics. (Summary by Alecia Biel) Plant Cell 10.1105/tpc.17.00452
Cation/H+ exchangers affect pollen wall formation, male fertility, and embryo development
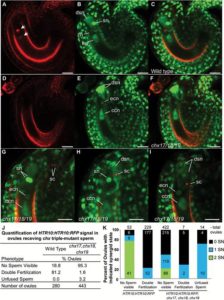 Cells have developed different mechanisms including control of ion and pH homeostasis to adapt to their constantly changing environment. Such adaption is accomplished by different ion transporters at the membranes. AtCHX17, AtCHX18 and AtCHX20 are members of the cation-H+ exchanger (CHX) family, which in Arabidopsis is formed by 28 members. Padmanaban et al. studied their function in planta. Previous studies showed the role of CHXs in K+ modulation and pH homeostasis by complementation of yeast mutants. The triple mutant chx17chx18chx19 showed decreased seed production and reciprocal crosses showed a pronounced male defect. Further analysis of the pods showed that there was pollen tube formation but fertilization did not occur. Moreover, live imaging of ovules pollinated with the chx triple mutant showed that only the egg or the central cell was fertilized but not both. Therefore loss of the CHX17, CHX18, and CHX19 genes lead to unsuccessful double fertilization. Scanning electron micrographs also revealed a disorganized pollen wall in the chx triple mutants compared to the wild type. The study shows the importance of the cation transporters during pollen development and sperm function. (Summary by Cecilia Vasquez-Robinet) J. Exp Bot. 10.1093/jxb/erw483
Cells have developed different mechanisms including control of ion and pH homeostasis to adapt to their constantly changing environment. Such adaption is accomplished by different ion transporters at the membranes. AtCHX17, AtCHX18 and AtCHX20 are members of the cation-H+ exchanger (CHX) family, which in Arabidopsis is formed by 28 members. Padmanaban et al. studied their function in planta. Previous studies showed the role of CHXs in K+ modulation and pH homeostasis by complementation of yeast mutants. The triple mutant chx17chx18chx19 showed decreased seed production and reciprocal crosses showed a pronounced male defect. Further analysis of the pods showed that there was pollen tube formation but fertilization did not occur. Moreover, live imaging of ovules pollinated with the chx triple mutant showed that only the egg or the central cell was fertilized but not both. Therefore loss of the CHX17, CHX18, and CHX19 genes lead to unsuccessful double fertilization. Scanning electron micrographs also revealed a disorganized pollen wall in the chx triple mutants compared to the wild type. The study shows the importance of the cation transporters during pollen development and sperm function. (Summary by Cecilia Vasquez-Robinet) J. Exp Bot. 10.1093/jxb/erw483
An acidophilic green algal genome provides insights into adaptation to an acidic environment ($)
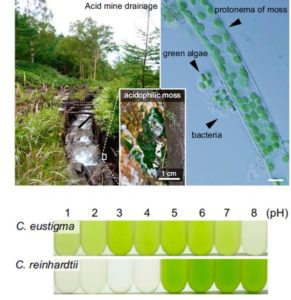 Hirooka et al. examined the genome of an acid-loving green alga, Chlamydomonas eustigma, to learn how it tolerates its low pH environment. Key differences between the acidophilic species and the neutrophilic species Chlamydomonas reinhardtii include: an increase in expression of genes encoding plasma membrane H+-ATPases (to better extrude protons) and heat-shock proteins (for molecular stress tolerance). Further, the acidophilic species has lost genes involved in organic acid-producing fermentation pathways, and has acquired genes involved in arsenic detoxification through horizontal gene transfer; arsenic is more readily taken up in acidic environments. This study extends our understanding of the requirements for life at low pH. The authors also suggest that acidophilic algae can be useful for food or fuel cultivation outdoors, as they can be grown in acidic conditions without risk of contamination by other organisms. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1707072114.
Hirooka et al. examined the genome of an acid-loving green alga, Chlamydomonas eustigma, to learn how it tolerates its low pH environment. Key differences between the acidophilic species and the neutrophilic species Chlamydomonas reinhardtii include: an increase in expression of genes encoding plasma membrane H+-ATPases (to better extrude protons) and heat-shock proteins (for molecular stress tolerance). Further, the acidophilic species has lost genes involved in organic acid-producing fermentation pathways, and has acquired genes involved in arsenic detoxification through horizontal gene transfer; arsenic is more readily taken up in acidic environments. This study extends our understanding of the requirements for life at low pH. The authors also suggest that acidophilic algae can be useful for food or fuel cultivation outdoors, as they can be grown in acidic conditions without risk of contamination by other organisms. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1707072114.
Tracking effector delivery in Irish famine potato pathogen
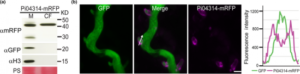 Pathogenic microbes interfere with the host cellular and physiological processes to promote infection. This interaction is monitored by pathogen molecules called effectors that either act in intercellular space or enter the host cells. Mechanisms underlying the uptake of these effectors are not fully understood. Oomycete effectors including domain-specific RXLR effectors have provided insights into effector biology over last decade. Even though oomycete effectors have been thoroughly studied as compared to fungal effectors, there has been no clear experimental evidence of their delivery through a particular infectious structure as was shown for effectors of the fungal pathogen M. oryzae several years ago. Now, Wang et al. have clearly shown experimental evidence of RXLR effector delivery through haustoria into plant cells. The authors have traced the localization of a predicted cytoplasmic effector by live-cell imaging using a transgenic strain of pathogen delivering fluorescent-tag fused effector as well as free GFP to track the hyphal growth. This has provided a clear experimental confirmation of RXLR effectors being delivered through haustoria. However, how the effector translocation step occurs still remains elusive and whether the RXLR motif drives translocation remains to be resolved. (Summary by Amey Redkar) New Phytol. 10.1111/nph.14696/full
Pathogenic microbes interfere with the host cellular and physiological processes to promote infection. This interaction is monitored by pathogen molecules called effectors that either act in intercellular space or enter the host cells. Mechanisms underlying the uptake of these effectors are not fully understood. Oomycete effectors including domain-specific RXLR effectors have provided insights into effector biology over last decade. Even though oomycete effectors have been thoroughly studied as compared to fungal effectors, there has been no clear experimental evidence of their delivery through a particular infectious structure as was shown for effectors of the fungal pathogen M. oryzae several years ago. Now, Wang et al. have clearly shown experimental evidence of RXLR effector delivery through haustoria into plant cells. The authors have traced the localization of a predicted cytoplasmic effector by live-cell imaging using a transgenic strain of pathogen delivering fluorescent-tag fused effector as well as free GFP to track the hyphal growth. This has provided a clear experimental confirmation of RXLR effectors being delivered through haustoria. However, how the effector translocation step occurs still remains elusive and whether the RXLR motif drives translocation remains to be resolved. (Summary by Amey Redkar) New Phytol. 10.1111/nph.14696/full
Special delivery: An independent secretion pathway for the delivery of cytoplasmic pathogen effectors
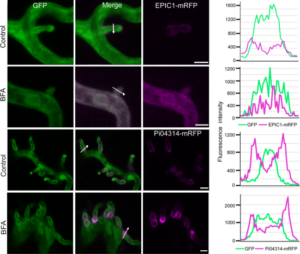 Pathogenic microbes manipulate host plants by secreting effector molecules that interfere with immunity. Bacterial phytopathogens achieve this using specialized secretion apparatuses that act as molecular ‘hypodermic needles’ to inject effector proteins directly into plant cells. In comparison, effector delivery in filamentous oomycete and fungal pathogens is less clear. Many filamentous pathogens develop specialized biotrophic structures like haustoria, which invaginate plant cells to deliver apoplastic and cytoplasmic effector molecules. In a recent study, Wang et al. examined microbial secretion pathways involved in the delivery of apoplastic and cytoplasmic effectors in the late blight pathogen Phytophthora infestans. By combining pharmacological treatments (brefeldin A) with live-cell imaging of pathogen effector proteins using fluorescent reporter strains, the authors demonstrate that apoplastic effectors are delivered by conventional secretion pathways whereas cytoplasmic effectors utilize an independent, BFA-insensitive route. These findings begin to tease apart the intricate processes involved in the delivery of effectors at the haustoria of filamentous pathogens. (Summary by Philip Carella) New Phytol. 10.1111/nph.14696/full
Pathogenic microbes manipulate host plants by secreting effector molecules that interfere with immunity. Bacterial phytopathogens achieve this using specialized secretion apparatuses that act as molecular ‘hypodermic needles’ to inject effector proteins directly into plant cells. In comparison, effector delivery in filamentous oomycete and fungal pathogens is less clear. Many filamentous pathogens develop specialized biotrophic structures like haustoria, which invaginate plant cells to deliver apoplastic and cytoplasmic effector molecules. In a recent study, Wang et al. examined microbial secretion pathways involved in the delivery of apoplastic and cytoplasmic effectors in the late blight pathogen Phytophthora infestans. By combining pharmacological treatments (brefeldin A) with live-cell imaging of pathogen effector proteins using fluorescent reporter strains, the authors demonstrate that apoplastic effectors are delivered by conventional secretion pathways whereas cytoplasmic effectors utilize an independent, BFA-insensitive route. These findings begin to tease apart the intricate processes involved in the delivery of effectors at the haustoria of filamentous pathogens. (Summary by Philip Carella) New Phytol. 10.1111/nph.14696/full
Root phonotropism: Early signalling events following sound perception in Arabidopsis roots
 Plants can hear. We know plants respond to touch, can perceive day and night, and respond to volatile compounds, water and nutrients. Moreno et al. studied root phonotropism (not phototropism). They showed that roots of Arabidopsis plants can perceive and respond to sound waves. Arabidopsis roots grew towards a sound source even in the absence of light, and sound caused an increase in cytosolic calcium. Continuous sound treatment for 9-10 days could reduce the K+ content in the roots, which was verified using electrophysiological measurements. Additionally, in Arabidopsis cells sound could increase superoxide production. The authors suggest that perception of sound is regulated by changes in Ca2+ and K+ ion fluxes with reactive oxygen species such as superoxide involved in signal amplification. This phenomenon throws light on how plants hear and perceive the sound of a distant water source in soil to direct roots to grow towards it. (Summary by Edna Anthony) Plant Science https://doi.org/10.1016/j.plantsci.2017.08.001
Plants can hear. We know plants respond to touch, can perceive day and night, and respond to volatile compounds, water and nutrients. Moreno et al. studied root phonotropism (not phototropism). They showed that roots of Arabidopsis plants can perceive and respond to sound waves. Arabidopsis roots grew towards a sound source even in the absence of light, and sound caused an increase in cytosolic calcium. Continuous sound treatment for 9-10 days could reduce the K+ content in the roots, which was verified using electrophysiological measurements. Additionally, in Arabidopsis cells sound could increase superoxide production. The authors suggest that perception of sound is regulated by changes in Ca2+ and K+ ion fluxes with reactive oxygen species such as superoxide involved in signal amplification. This phenomenon throws light on how plants hear and perceive the sound of a distant water source in soil to direct roots to grow towards it. (Summary by Edna Anthony) Plant Science https://doi.org/10.1016/j.plantsci.2017.08.001
Stem parasitic plant Cuscuta australis (dodder) transfers herbivory-induced signals among plants
 Dodders (Cuscuta spp) are parasitic plants, which absorb water and nutrients from their host. Their vines can embrace, and in this way connect, more than one host. Hettenhausen et al. showed that in certain situations these connections serve as communication routes. In their experiment a pair of soybean plants were parasitized by Cuscuta australis and thereafter one plant from each pair was subjected to caterpillar attack. The herbivore action led to increase in the expression of various defense related genes. The response was observed not only in caterpillar-attacked plants but also in the undamaged ones. Importantly the previously undamaged plants showed higher resistance to subsequent herbivore treatment. The results indicate that the herbivory-induced signal was transmitted by Cuscuta australis vines leading to the activation of defense mechanisms. The study reveals new aspects of signaling and communication and shows that in certain conditions plants can benefit even from parasitic infection. (Summary by Jagna Chmielowska-Bąk) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1704536114
Dodders (Cuscuta spp) are parasitic plants, which absorb water and nutrients from their host. Their vines can embrace, and in this way connect, more than one host. Hettenhausen et al. showed that in certain situations these connections serve as communication routes. In their experiment a pair of soybean plants were parasitized by Cuscuta australis and thereafter one plant from each pair was subjected to caterpillar attack. The herbivore action led to increase in the expression of various defense related genes. The response was observed not only in caterpillar-attacked plants but also in the undamaged ones. Importantly the previously undamaged plants showed higher resistance to subsequent herbivore treatment. The results indicate that the herbivory-induced signal was transmitted by Cuscuta australis vines leading to the activation of defense mechanisms. The study reveals new aspects of signaling and communication and shows that in certain conditions plants can benefit even from parasitic infection. (Summary by Jagna Chmielowska-Bąk) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1704536114
Less is more: Gene loss in flower pollination evolution ($)
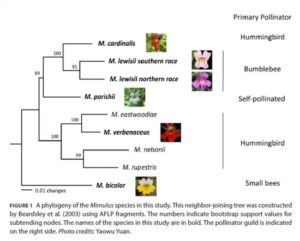 The evolution of flowers solved one of the largest obstacles of plant reproduction, finding a compatible mate. Since plants are sedentary, they are unable to search for a compatible mate like other organisms. Instead they use pollinators to do the searching for them. Flowers use scent and color to attract pollinators, and many offer rewards of nectar and pollen for the work they do. Peng and others investigated the floral biology of several monkeyflower species. They found that although monkeyflowers all evolved from a common ancestor that was insect-pollinated, many species have evolved other pollination strategies like small-bee, bumblebee, hummingbird, and self pollination. They found that the gene that controls the scent used to attract bumblebees in Lewis monkeyflower, the OCIMENE SYNTHASE (OS) gene, has had several independent loss-of-function mutations. These mutations led to changes in scent type and quantity, affecting which pollinators are attracted. Mutations of the OS gene may have been involved in the process of reproductive isolation by attracting different pollinators. Although other genetic mutations, like the development of red pigments to attract hummingbirds, are also involved in pollination strategies, this research shows how vital genetic mutations are for pollinator attraction and species evolution. (Summary by Jason Stettler) Am. J. Bot. 10.3732/ajb.1700104
The evolution of flowers solved one of the largest obstacles of plant reproduction, finding a compatible mate. Since plants are sedentary, they are unable to search for a compatible mate like other organisms. Instead they use pollinators to do the searching for them. Flowers use scent and color to attract pollinators, and many offer rewards of nectar and pollen for the work they do. Peng and others investigated the floral biology of several monkeyflower species. They found that although monkeyflowers all evolved from a common ancestor that was insect-pollinated, many species have evolved other pollination strategies like small-bee, bumblebee, hummingbird, and self pollination. They found that the gene that controls the scent used to attract bumblebees in Lewis monkeyflower, the OCIMENE SYNTHASE (OS) gene, has had several independent loss-of-function mutations. These mutations led to changes in scent type and quantity, affecting which pollinators are attracted. Mutations of the OS gene may have been involved in the process of reproductive isolation by attracting different pollinators. Although other genetic mutations, like the development of red pigments to attract hummingbirds, are also involved in pollination strategies, this research shows how vital genetic mutations are for pollinator attraction and species evolution. (Summary by Jason Stettler) Am. J. Bot. 10.3732/ajb.1700104