What We’re Reading: September 1st
This week’s issue of What We’re Reading is guest edited by Arif Ashraf, a PhD student at Iwate University, Japan, and Graduate Student Ambassador of ASPB. His research interest is understanding the hormonal interplay in primary root development of Arabidopsis thaliana. He blogs about plant science at http://www.aribidopsis.com/. Arif is available in twitter (@aribidopsis) and Google plus (+Arif). Let’s see what Arif and friends chose to feature this week:
SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis ($)
 Light is one of the environmental conditions which regulates plants’ development, but knowledge of how light-induced transcript accumulation occurs via transcriptional versus post-transcriptional mechanisms is limited. Xin et al. report Splicing Factor for Phytochrome Signaling (SFPS) as an interacting partner of the photoreceptor Phytochrome B. The sfps mutant has a hyposensitive response to light and controls pre-mRNA processing of myriad genes involved in light signaling, photosynthesis, and the circadian clock. Additionally, the study of the quadruple pif mutant (pifq) revealed that Phytochrome Interacting Factors (PIFs) work downstream of SFPS. The authors propose a model where light-dependent development is regulated via SFPS, by controlling pre-mRNA processing of light signaling and circadian clock genes. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1706379114
Light is one of the environmental conditions which regulates plants’ development, but knowledge of how light-induced transcript accumulation occurs via transcriptional versus post-transcriptional mechanisms is limited. Xin et al. report Splicing Factor for Phytochrome Signaling (SFPS) as an interacting partner of the photoreceptor Phytochrome B. The sfps mutant has a hyposensitive response to light and controls pre-mRNA processing of myriad genes involved in light signaling, photosynthesis, and the circadian clock. Additionally, the study of the quadruple pif mutant (pifq) revealed that Phytochrome Interacting Factors (PIFs) work downstream of SFPS. The authors propose a model where light-dependent development is regulated via SFPS, by controlling pre-mRNA processing of light signaling and circadian clock genes. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1706379114
Establishment of photosynthesis is controlled by two distinct regulatory phases ($)
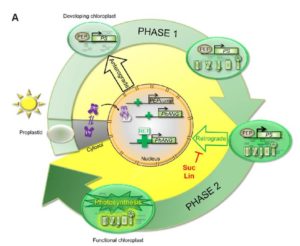 Chloroplast biogenesis and leaf development are closely linked processes that have been difficult to tease apart, until recently. Dubreuil et al. used a pluripotent inducible cell line from Arabidopsis, basically plant stem cells, which develop chloroplasts on demand when exposed to light. Using this system as well as developing maize leaves, the researchers proposed a two-phase system for chloroplast development. In phase one, there is a rapid change in nuclear gene expression (3084 differentially expressed genes between Zero and One day of light), accompanied by a shift in the cells’ metabolome. Between day Four and Five, the second phase begins with another nuclear gene expression transition (12,411 differentially expressed genes), including chloroplast and chromatin modification genes. This change was correlated with the appearance of intact and functional photosynthetic units within the thylakoid membranes. Chloroplasts also changed shape from tubes to globular organelles and started to move away from the nucleus towards the cell cortex. The second phase could be inhibited by sucrose or lincomycin suggesting that it depends on retrograde signaling. This paper is a fantastic start to the full dissection of chloroplast biogenesis process. Plant Physiol. 10.1104/pp.17.00435 (Contributed by Jennifer Robinson)
Chloroplast biogenesis and leaf development are closely linked processes that have been difficult to tease apart, until recently. Dubreuil et al. used a pluripotent inducible cell line from Arabidopsis, basically plant stem cells, which develop chloroplasts on demand when exposed to light. Using this system as well as developing maize leaves, the researchers proposed a two-phase system for chloroplast development. In phase one, there is a rapid change in nuclear gene expression (3084 differentially expressed genes between Zero and One day of light), accompanied by a shift in the cells’ metabolome. Between day Four and Five, the second phase begins with another nuclear gene expression transition (12,411 differentially expressed genes), including chloroplast and chromatin modification genes. This change was correlated with the appearance of intact and functional photosynthetic units within the thylakoid membranes. Chloroplasts also changed shape from tubes to globular organelles and started to move away from the nucleus towards the cell cortex. The second phase could be inhibited by sucrose or lincomycin suggesting that it depends on retrograde signaling. This paper is a fantastic start to the full dissection of chloroplast biogenesis process. Plant Physiol. 10.1104/pp.17.00435 (Contributed by Jennifer Robinson)
ePlant: Visualizing and exploring multiple levels of data for hypothesis generation ($)
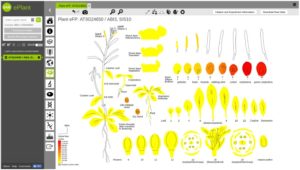 The application of systems biology is quite phenomenal these days for prediction-based modeling and interactive data visualization. Along with the genome sequencing of the model plant Arabidopsis thaliana, there has been a parallel increase in systems biology tools. Unfortunately, these tools have been developed by different groups or institutions for various purposes, whereas a unifying and comprehensive systems biology toolbox is more convenient to connect multiple databases and generate prediction models or build hypothesis. ePlant provides that purpose for Arabidopsis thaliana. In ePlant, Wrase et al. incorporated multiple datasets and integrated search system for analysis and visualization in a single portal. Here, users may access genome, proteome, interactome, transcriptome, sub-cellular localization, ecotype distribution, and 3D molecular structure data. ePlant provides lucid visualization for multiple level of data in a single platform, and can be built upon for research in other species. Plant Cell 10.1105/tpc.17.00073
The application of systems biology is quite phenomenal these days for prediction-based modeling and interactive data visualization. Along with the genome sequencing of the model plant Arabidopsis thaliana, there has been a parallel increase in systems biology tools. Unfortunately, these tools have been developed by different groups or institutions for various purposes, whereas a unifying and comprehensive systems biology toolbox is more convenient to connect multiple databases and generate prediction models or build hypothesis. ePlant provides that purpose for Arabidopsis thaliana. In ePlant, Wrase et al. incorporated multiple datasets and integrated search system for analysis and visualization in a single portal. Here, users may access genome, proteome, interactome, transcriptome, sub-cellular localization, ecotype distribution, and 3D molecular structure data. ePlant provides lucid visualization for multiple level of data in a single platform, and can be built upon for research in other species. Plant Cell 10.1105/tpc.17.00073
The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants
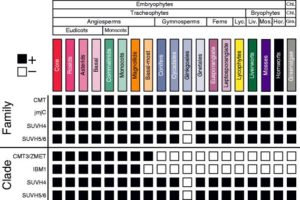 In land plant genomes, transposable elements (TEs) are ubiquitously methylated in CG and non-CG sequence contexts. Apart from methylation of TEs, DNA methylation also occurs on bodies of actively transcribed genes, typically in the CG context, with very low or no non-CG methylation, and is referred to as gene body methylation (gbM). To begin to understand the evolution, origins, and functions of gbM and its relationship to CHROMOMETHYLTRANSFERASE (CMT) proteins, Bewick et al., used data from more than 1000 transcriptomes and 77 methylomes for phylogenetic and methylation variation analysis. Their extensive taxonomic sampling redefined the CMT clade and showed that CMT genes originated prior to the evolution of land plants. CMT3 has been shown to maintain methylation at CHG sites, and CMT2 methylates CHH sites in the heterochromatin, while CMT1 is non-functional. Apart from maintaining non-CG methylation at TEs in the pericentromeric regions, CMTs are also important for CG methylation within gbM genes. In Brassicaceae, particularly in the clade containing B. oleracea, B. rapa, and S. parvula, CMT3 has a higher rate of molecular evolution measured by dN/dS compared to other eudicots, potentially disrupting CMT3 function to varying degrees. The authors also show the tightly correlated evolution of CMT3 and IBM1, a histone demethylase which preferentially demethylates at H3K9, suggesting their involvement in the gbM pathway. These results open avenues for future work to help better understand the consequences of and the relationship between DNA gene body methylation and gene expression. Genome Biol. 10.1186/s13059-017-1195-1 (Contributed by Sunil Kumar K R)
In land plant genomes, transposable elements (TEs) are ubiquitously methylated in CG and non-CG sequence contexts. Apart from methylation of TEs, DNA methylation also occurs on bodies of actively transcribed genes, typically in the CG context, with very low or no non-CG methylation, and is referred to as gene body methylation (gbM). To begin to understand the evolution, origins, and functions of gbM and its relationship to CHROMOMETHYLTRANSFERASE (CMT) proteins, Bewick et al., used data from more than 1000 transcriptomes and 77 methylomes for phylogenetic and methylation variation analysis. Their extensive taxonomic sampling redefined the CMT clade and showed that CMT genes originated prior to the evolution of land plants. CMT3 has been shown to maintain methylation at CHG sites, and CMT2 methylates CHH sites in the heterochromatin, while CMT1 is non-functional. Apart from maintaining non-CG methylation at TEs in the pericentromeric regions, CMTs are also important for CG methylation within gbM genes. In Brassicaceae, particularly in the clade containing B. oleracea, B. rapa, and S. parvula, CMT3 has a higher rate of molecular evolution measured by dN/dS compared to other eudicots, potentially disrupting CMT3 function to varying degrees. The authors also show the tightly correlated evolution of CMT3 and IBM1, a histone demethylase which preferentially demethylates at H3K9, suggesting their involvement in the gbM pathway. These results open avenues for future work to help better understand the consequences of and the relationship between DNA gene body methylation and gene expression. Genome Biol. 10.1186/s13059-017-1195-1 (Contributed by Sunil Kumar K R)
Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature
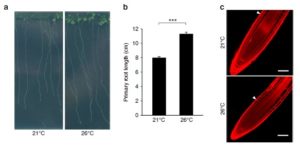 Temperature is one of the major factors affecting plant growth and development. Martins et al. showed that elevated temperature increases root growth through elongated cell size, but reduced both meristem size and number of meristematic cells. The elongation of root growth during elevated temperature is independent of auxin and confirmed through the study of auxin mutants, response markers, and whole genome-wide mRNA-seq analysis. In contrast, the brassinosteroid signaling component BRI1 acts as a negative regulator during high temperature-mediated root growth elongation. Interestingly, high temperature doesn’t alter BRI1 transcripts level, but downregulates in protein level. The down-regulation of BRI1 in high temperature in controlled through post-translational modification, more precisely ubiquitination. Nature Comms. 10.1038/s41467-017-00355-4
Temperature is one of the major factors affecting plant growth and development. Martins et al. showed that elevated temperature increases root growth through elongated cell size, but reduced both meristem size and number of meristematic cells. The elongation of root growth during elevated temperature is independent of auxin and confirmed through the study of auxin mutants, response markers, and whole genome-wide mRNA-seq analysis. In contrast, the brassinosteroid signaling component BRI1 acts as a negative regulator during high temperature-mediated root growth elongation. Interestingly, high temperature doesn’t alter BRI1 transcripts level, but downregulates in protein level. The down-regulation of BRI1 in high temperature in controlled through post-translational modification, more precisely ubiquitination. Nature Comms. 10.1038/s41467-017-00355-4
POSITIVE REGULATOR OF IRON HOMEOSTASIS 1, OsPRI1, facilitates iron homeostasis ($)
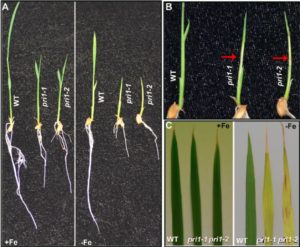 Zhang et al. showed that POSITIVE REGULATOR OF IRON HOMEOSTASIS 1 (OsPRI1), a bHLH transcription factor, is an interacting partner of the iron-binding sensor OsHRZ1. A loss-of-function mutation of OsPRI1 is responsible for a hypersensitive response to Fe deficiency. OsPRI1 works downstream of OsHRZ1 and directly controls the expression of OsIRO2 and OsIRO3 by binding to the promoter regions. At the same time, the stability of OsPRI1 was negatively correlated with OsHRZ1 production, because OsPRI1 is subjected to ubiquitination by OsHRZ1. This study has provided a very important missing link (OsPRI1) between the iron-binding sensor OsHRZ1 and the Fe-deficiency–responsive genes OsIRO2 and OsIRO3 to define the OsHRZ1–OsPRI1–OsIRO2/3 signal transduction cascade in rice. Plant Physiol. 10.1104/pp.17.00794
Zhang et al. showed that POSITIVE REGULATOR OF IRON HOMEOSTASIS 1 (OsPRI1), a bHLH transcription factor, is an interacting partner of the iron-binding sensor OsHRZ1. A loss-of-function mutation of OsPRI1 is responsible for a hypersensitive response to Fe deficiency. OsPRI1 works downstream of OsHRZ1 and directly controls the expression of OsIRO2 and OsIRO3 by binding to the promoter regions. At the same time, the stability of OsPRI1 was negatively correlated with OsHRZ1 production, because OsPRI1 is subjected to ubiquitination by OsHRZ1. This study has provided a very important missing link (OsPRI1) between the iron-binding sensor OsHRZ1 and the Fe-deficiency–responsive genes OsIRO2 and OsIRO3 to define the OsHRZ1–OsPRI1–OsIRO2/3 signal transduction cascade in rice. Plant Physiol. 10.1104/pp.17.00794
The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana
 The histone H3 variant H3.3 is distinguished by its expression throughout the cell cycle, while H3.1 is expressed predominantly during DNA replication. Genome-wide ChIP studies in plants have shown that H3.3 is associated with actively transcribed genes, and enriched near the transcriptional end sites. This pattern also overlaps with the enrichment of RNA Polymerase II. DNA methylation frequently occurs over bodies of actively transcribed genes in animals and vascular plants. In Arabidopsis, profiles of gene body methylation overlap with enrichment for H3.3. To understand the mechanism involved, Wollmann et al., engineered H3.3 knock down in A. thaliana. They observed a reduction in transcription of environment responsive genes and transcription-dependent increase in linker histone H1 distribution over gene bodies. Their data also suggest that H3K36 methylation and other transcription-related H3 modifications do not seem to play any role in gene body methylation. Finally, the authors propose a mechanism wherein H3.3 prevents the recruitment of H1, which in turn inhibits H1’s promotion of chromatin folding to restrict access to DNA methyltransferases. Thus, H3.3 helps provide access to methyltransferases and likely shapes transcription-dependent gene body methylation patterns in Arabidopsis. Genome Biol. 10.1186/s13059-017-1221-3 (Contributed by Sunil Kumar K R)
The histone H3 variant H3.3 is distinguished by its expression throughout the cell cycle, while H3.1 is expressed predominantly during DNA replication. Genome-wide ChIP studies in plants have shown that H3.3 is associated with actively transcribed genes, and enriched near the transcriptional end sites. This pattern also overlaps with the enrichment of RNA Polymerase II. DNA methylation frequently occurs over bodies of actively transcribed genes in animals and vascular plants. In Arabidopsis, profiles of gene body methylation overlap with enrichment for H3.3. To understand the mechanism involved, Wollmann et al., engineered H3.3 knock down in A. thaliana. They observed a reduction in transcription of environment responsive genes and transcription-dependent increase in linker histone H1 distribution over gene bodies. Their data also suggest that H3K36 methylation and other transcription-related H3 modifications do not seem to play any role in gene body methylation. Finally, the authors propose a mechanism wherein H3.3 prevents the recruitment of H1, which in turn inhibits H1’s promotion of chromatin folding to restrict access to DNA methyltransferases. Thus, H3.3 helps provide access to methyltransferases and likely shapes transcription-dependent gene body methylation patterns in Arabidopsis. Genome Biol. 10.1186/s13059-017-1221-3 (Contributed by Sunil Kumar K R)
In Arabidopsis thaliana cadmium affects growth of the primary root by altering SCR expression and auxin-cytokinin crosstalk
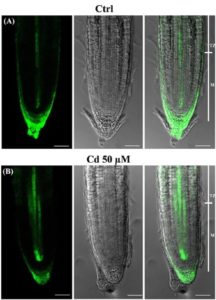 Cadmium is a toxic metal in our environment. Bruno et al. explored cadmium-mediated root-growth inhibition using the model plant Arabidopsis thaliana. Cadmium affects root growth longitudinally by reducing root meristem cell number and radially by controlling the number and width of stele cells. The authors showed that misexpression of the trasncription factor SCARECROW (SCR) occurs due to cadmium toxicity in roots. Two major hormones – auxin and cytokinin – are crucial for root developmental processes. Studies with auxin responsive marker (pDR5::GFP), auxin transporters (PIN1, 2, 3 and 7) and cytokinin response marker (TCSn::GFP) revealed that cadmium induces down-regulation and mislocalization of PIN proteins as well as altered auxin maxima in the root tip. Furthermore, an increased signal of TCSn::GFP is detected due to cadmium. This work provides a comprehensive overview of how cadmium alters root development from the physiological and molecular perspectives. Front. Plant Sci. 10.3389/fpls.2017.01323
Cadmium is a toxic metal in our environment. Bruno et al. explored cadmium-mediated root-growth inhibition using the model plant Arabidopsis thaliana. Cadmium affects root growth longitudinally by reducing root meristem cell number and radially by controlling the number and width of stele cells. The authors showed that misexpression of the trasncription factor SCARECROW (SCR) occurs due to cadmium toxicity in roots. Two major hormones – auxin and cytokinin – are crucial for root developmental processes. Studies with auxin responsive marker (pDR5::GFP), auxin transporters (PIN1, 2, 3 and 7) and cytokinin response marker (TCSn::GFP) revealed that cadmium induces down-regulation and mislocalization of PIN proteins as well as altered auxin maxima in the root tip. Furthermore, an increased signal of TCSn::GFP is detected due to cadmium. This work provides a comprehensive overview of how cadmium alters root development from the physiological and molecular perspectives. Front. Plant Sci. 10.3389/fpls.2017.01323
VPS9a activates the Rab5 GTPase ARA7 to confer distinct pre- and post-invasive plant innate immunity ($)
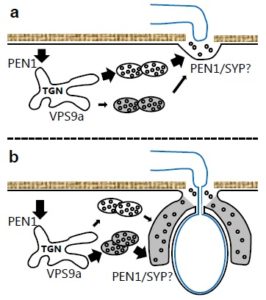
Fungal invasion exploits plant cell membrane trafficking components. For instance, plant immunity against fungi such as Vlumeria graminis f.sp. hordei (Bgh) is defined through two stages: pre- and post-invasive immunity. Nielsen et al. found that Vacuolar Protein Sorting 9a (VPS9a) acts as a guanine nucleotide exchange factor which activates the Rab5 GTPase family (ARA6, ARA7 and RHA1). Formation of Multi Vesicular Bodies (MVBs) and proper membrane targeting depend on Rab5 GTPases. This article sheds light on the role of VPS9a for both pre- and post-invasive immunity against Bgh and broadens our understanding about membrane trafficking during powdery mildew attack. The authors conclude, “We propose that VPS9a plays a conserved role in organizing cellular endomembrane trafficking, required for delivery of defense components in response to powdery mildew fungi.” Plant Cell 10.1105/tpc.16.00859
The ARM domain of ARMADILLO-REPEAT KINESIS 1 is not required for microtubule catastrophe but can negatively regulate NIMA-RELATED KINASE 6 in Arabidopsis thaliana
 ARMADILLO-REPEAT KINESIN 1 (ARK1) promotes microtubule disassembly and NIMA-RELATED KINASE 6 (NEK6) organizes microtubule arrays. Previous studies showed that NEK6 interacts with ARK1 through Armadillo-repeat (ARM) cargo domain. Eng et al. looked at two constructs: one which lacks ARM (ARK1▲ARM-GFP) and another one with only the ARM domain (ARM-GFP). Interestingly, the authors have found that ARK1▲ARM-GFP can induce microtubule disassenbly and rescue the ark1-1 root hair phenotype. This indicates that ARK1 doesn’t require the ARM domain or interaction with NEK6 to induce microtubule catastrophe. The authors conclude that ARK1 and NEK6 contribute to microtubule dynamics independently, and observe that NEK6 gene expression is blocked when ARK’s ARM domain is overexpressed, suggesting a relationship at the level of transcriptional regulation. This study was featured on the cover of Plant & Cell Physiology (Volume 58, Issue 8, August 2017). Plant Cell Physiol. 10.1093/pcp/pcx070
ARMADILLO-REPEAT KINESIN 1 (ARK1) promotes microtubule disassembly and NIMA-RELATED KINASE 6 (NEK6) organizes microtubule arrays. Previous studies showed that NEK6 interacts with ARK1 through Armadillo-repeat (ARM) cargo domain. Eng et al. looked at two constructs: one which lacks ARM (ARK1▲ARM-GFP) and another one with only the ARM domain (ARM-GFP). Interestingly, the authors have found that ARK1▲ARM-GFP can induce microtubule disassenbly and rescue the ark1-1 root hair phenotype. This indicates that ARK1 doesn’t require the ARM domain or interaction with NEK6 to induce microtubule catastrophe. The authors conclude that ARK1 and NEK6 contribute to microtubule dynamics independently, and observe that NEK6 gene expression is blocked when ARK’s ARM domain is overexpressed, suggesting a relationship at the level of transcriptional regulation. This study was featured on the cover of Plant & Cell Physiology (Volume 58, Issue 8, August 2017). Plant Cell Physiol. 10.1093/pcp/pcx070
The chickpea Early Flowering 1 (Efl1) locus is an ortholog of Arabidopsis ELF3 ($)
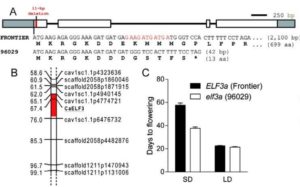 Chickpea is a highly cultivated member of the legume family, mostly in India, Australia, Pakistan, Turkey, Burma, Iran, Canada and the US. The detrimental threat for chickpea is the invasion of Ascochyta blight, which is caused by the fungal pathogen Ascochyta rabiei (formerly known as Phoma rabiei). The two possible ways to combat this problem are – finding resistance genes or finding early flowering genes. Ridge et al. took the latter approach. They mapped, sequenced and characterized an early flowering gene, Efl1, in chickpea. Interestingly, Efl1 is an ortholog of the Arabidopsis early flowering gene EFL3. The authors identified an 11-bp deletion in the first exon which causes premature termination in the early-flowering variety. A search through 236 lines from global germplasm resources confirmed that 13 lines contain this 11-bp deletion. The variation has arisen in recent times within Indian germplasm to enhance an existing early flowering phenotype. Plant Physiol. 10.1104/pp.17.00082
Chickpea is a highly cultivated member of the legume family, mostly in India, Australia, Pakistan, Turkey, Burma, Iran, Canada and the US. The detrimental threat for chickpea is the invasion of Ascochyta blight, which is caused by the fungal pathogen Ascochyta rabiei (formerly known as Phoma rabiei). The two possible ways to combat this problem are – finding resistance genes or finding early flowering genes. Ridge et al. took the latter approach. They mapped, sequenced and characterized an early flowering gene, Efl1, in chickpea. Interestingly, Efl1 is an ortholog of the Arabidopsis early flowering gene EFL3. The authors identified an 11-bp deletion in the first exon which causes premature termination in the early-flowering variety. A search through 236 lines from global germplasm resources confirmed that 13 lines contain this 11-bp deletion. The variation has arisen in recent times within Indian germplasm to enhance an existing early flowering phenotype. Plant Physiol. 10.1104/pp.17.00082
Production of low-Cs+ rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR-Cas system
 Nuclear accidents in recent years such as the Fukushima incident during the tsunami in 2011 revealed the detrimental effects of leaked radioactive cesium (Cs) in environmental soil and water. Due to Cs’s chemical similarity with potassium, an essential macronutrient for plants, cesium is taken up by potassium transporters even when it is present in small amounts. Consequently, Cs can make its way to the edible parts of plants there are destined for the food chain. In this study, Nieves-Cordones et al. generated knock-out mutants the of potassium transporter OsHAK1 using the CRISPR-Cas system. Knock-out mutants show dramatically reduced uptake of cesium. In addition to lab experiments, the authors examined these knock-out mutants grown in radio-cesium contaminated soils and found a similarly reduced uptake of cesium. This paper shows a very practical approach to avoid cesium contamination in rice plants. Plant J. 10.1111/tpj.13632
Nuclear accidents in recent years such as the Fukushima incident during the tsunami in 2011 revealed the detrimental effects of leaked radioactive cesium (Cs) in environmental soil and water. Due to Cs’s chemical similarity with potassium, an essential macronutrient for plants, cesium is taken up by potassium transporters even when it is present in small amounts. Consequently, Cs can make its way to the edible parts of plants there are destined for the food chain. In this study, Nieves-Cordones et al. generated knock-out mutants the of potassium transporter OsHAK1 using the CRISPR-Cas system. Knock-out mutants show dramatically reduced uptake of cesium. In addition to lab experiments, the authors examined these knock-out mutants grown in radio-cesium contaminated soils and found a similarly reduced uptake of cesium. This paper shows a very practical approach to avoid cesium contamination in rice plants. Plant J. 10.1111/tpj.13632
Field-based species identification of closely-related plants using real-time nanopore sequencing
 DNA sequencing was slow before the development of high throughput sequencing. Portable DNA sequencing, which would make sequencing on-site a reality, was impossible until recently. Parker et al. report on the on-site use of MinION from Oxford Nanopore Technologies for DNA barcoding, which yields data within hours, showing “species identification could be conducted using genome scale data generated at the point of sample collection.” The paper describes the procedures that were followed to identify and distinguish samples from Arabidopsis thaliana and Arabidopsis lyrata, two closely related species. Their results demonstrate that “the entire process (from sample collection to genome scale sequencing) is now feasible for eukaryotic species within a few hours in field conditions.” Even with the caveats of lower sample purity and sequencing coverage, their data were useful for genome assembly and would also allow quick evolutionary analyses. As sequencing technology advances, it will be more and more reachable, even for non-experts, incredibly extending its uses—this is a nice time to do field work. Scientific Reports: 10.1038/s41598-017-08461-5 (Contributed by Gabriela Auge)
DNA sequencing was slow before the development of high throughput sequencing. Portable DNA sequencing, which would make sequencing on-site a reality, was impossible until recently. Parker et al. report on the on-site use of MinION from Oxford Nanopore Technologies for DNA barcoding, which yields data within hours, showing “species identification could be conducted using genome scale data generated at the point of sample collection.” The paper describes the procedures that were followed to identify and distinguish samples from Arabidopsis thaliana and Arabidopsis lyrata, two closely related species. Their results demonstrate that “the entire process (from sample collection to genome scale sequencing) is now feasible for eukaryotic species within a few hours in field conditions.” Even with the caveats of lower sample purity and sequencing coverage, their data were useful for genome assembly and would also allow quick evolutionary analyses. As sequencing technology advances, it will be more and more reachable, even for non-experts, incredibly extending its uses—this is a nice time to do field work. Scientific Reports: 10.1038/s41598-017-08461-5 (Contributed by Gabriela Auge)



