Save Time and Fish for the Clock
Plants live by the clock. It helps them to predict day and night as well as upcoming seasons, and to decide when it’s time for reproduction. These predictions depend on oscillating processes that involve gene transcription and protein stability, cycling in a period of, for example, 24 h. Environmental factors such as light, temperature, and humidity serve as the input to fine-tune these oscillations and enable the clock to adjust when needed. As output, a wide range of physiological and developmental traits optimize survival in the expected conditions. For instance, the clock gates end-of-night growth in Arabidopsis (Arabidopsis thaliana) seedlings (Martín et al., 2018), times heliotropism to make sunflowers (Helianthus annuus) face the sun at dawn (Atamian et al., 2016), optimizes the time of flowering within a season (Suárez-López et al., 2001), and fine-tunes lateral root outgrowth (Voß et al., 2015).
The function of the plant circadian clock depends on ubiquitination and degradation of proteins in recurring cycles. The “bridging” between the E3 ubiquitin ligase complex and these clock-related substrate proteins is facilitated by a small family of F-box proteins, consisting of ZEITLUPE (ZTL), LOV KELCH PROTEIN2 (LKP2), and FLAVIN-BINDING KELCH REPEAT F-BOX1 (FKF1). In this issue of Plant Physiology, Chin-Mei Lee et al. (2018) use a clever technique to tackle two major questions that could not have been answered using traditional genetic approaches: (1) Do these F-box proteins have ubiquitylation-independent functions? (2) Which substrate proteins are bound by ZTL1, LKP1, and FKF1?
Their method can be compared to fishing. It tricks proteins into binding a nonbridging (F-box domain-free) version of ZTL, LKP2, or FKF1. These “decoys” are dominant-negative, truncated forms of ZTL, LKP2, and FKF1 (simplified representation in Fig. 1). The dominance of the decoys over the full-length versions enabled the authors to study the role of the non-F-box domains of these proteins. They discovered that, whereas the clock period is affected by all three proteins in a ubiquitylation-dependent fashion, ZTL and LKP2 also can regulate flowering time independent of their F-box domain.
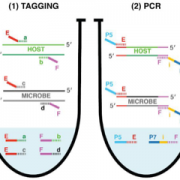
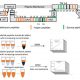
A simplified model of the decoy method described by Lee et al. (2018). To identify ubiquitylation-independent functions of F-box proteins and interacting substrates, a decoy protein, which lacks the F-box domain and is tagged with a marker, is overexpressed in wild-type Arabidopsis plants (1). The dominant negative proteins bind the substrate proteins (2), which results in phenotypes that can be linked to the F-box or other protein domains (3.1). Fishing with the decoy proteins by immunoprecipitation with anti-tag antibodies (2), followed by mass spectrometry, identifies their targets (3.2).
 Too
TooWhen the authors used the decoys for the real “fishing” experiment, immunoprecipitation followed by mass spectrometry (Fig. 1), they identified several known and new interactors of these clock regulators, without losing the substrates to the E3 ubiquitin-ligase pathways.
The decoy method shows great potential for applications in plant studies. Chin-Mei Lee and colleagues stress that it can be similarly applied to find substrates of other F-box proteins. As F-box proteins form a large family of approximately 700 members (Gagne et al., 2002), this opens doors in many different plant science research areas. And because of the dominant-negative approach, the decoy method tackles redundancy in larger subfamilies of F-box proteins and even genetic copies of genes encoding them.
In conclusion, Chin-Mei Lee et al. present a game-changing technique that allowed them to fish for the clock and will help others to save time.
REFERENCES
Atamian HS, Creux NM, Brown EA, Garner AG, Blackman BK, Harmer SL (2016) Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science353: 587–590
Gagne JM, Downes BP, Shiu S-H, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524
Martín G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CMM, Boix M, Henriques R, Minguet EG, Alabadí D, et al. (2018) Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr Biol 28: 311–318.e5
Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120
Voß U, Wilson MH, Kenobi K, Gould PD, Robertson FC, Peer WA, Lucas M, Swarup K, Casimiro I, Holman TJ, et al. (2015) The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat Commun 6: 7641