Managing Competing Interests: Partitioning S between Glutathione and Protein Synthesis
Sulfur (S) is an essential element for cell function and responses to the environment. The primary S source is sulfate, which, following uptake by specific transporters, is reduced and incorporated into the amino acids Cysteine (Cys) and Methionine, and thereafter into proteins and peptides, including the multifunctional tripeptide thiol glutathione (Takahashi et al., 2011). Cys production requires production of sulfide by sulfite reductase (SiR; Khan et al., 2010), followed by incorporation of sulfide into carbon skeletons to generate Cys via the Cys synthase complex in which Serine (Ser) acetyl transferase and O-acetyl-Ser(thiol)lyase cooperate to allow metabolite channeling (Fig. 1).
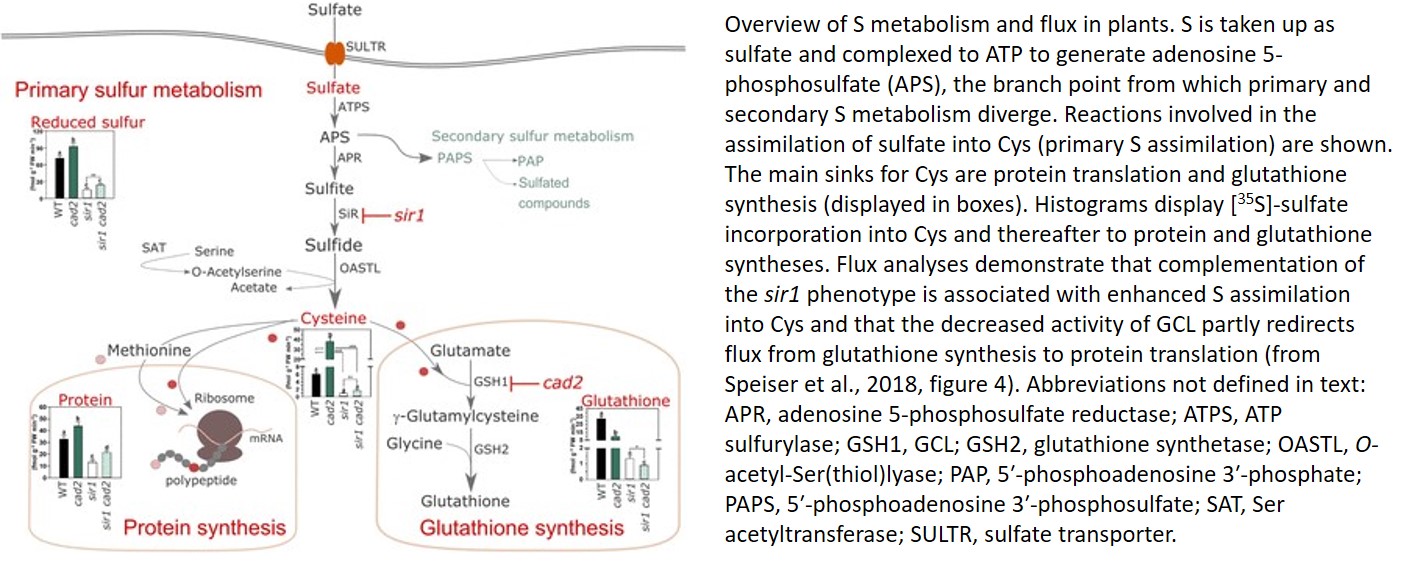
In plants, Cys biosynthesis is the unique entry point of reduced S into metabolism and is therefore key to coordinating the flux of S with those of carbon and nitrogen (Takahashi et al., 2011). Thus, sulfate assimilation is tightly regulated, in particular because downstream pathways such as protein translation and glutathione synthesis compete for the same source, Cys. Plants sense the availability of Cys precursors rather than Cys itself (Dong et al., 2017). In S-rich plants such as Arabidopsis thaliana, mechanisms regulating S assimilation are complex and far from fully understood (Takahashi et al., 2011). Regulatory nodes upstream of Cys involve intermediate metabolites (e.g. O-acetyl-Ser) and both transcriptional and posttranscriptional control at the level of sulfate transporters, adenosine phosphosulfate reductases, and Ser acetyl transferases (Bick et al., 2001; Hopkins et al., 2005; Queval et al., 2009; Takahashi et al., 2011). Yet, the factors that determine S distribution downstream of Cys are unclear.
In this issue of Plant Physiology, Speiser et al. (2018) identify the activity of the first committed enzyme of glutathione synthesis, Glutamate Cys ligase (GCL/γ-ECS/CAD2), as a key determinant of S flux into not only glutathione but also protein synthesis. The authors show that the drastic growth arrest of the sir1 mutant is partially rescued when glutathione deficiency is introduced via the cad2 mutation, which decreases glutathione levels to 30% to 40% of wild-type levels (Cobbett et al., 1998), or pharmacologically through treatment with the GCL inhibitor buthionine sulfoximine. Interestingly, however, it is unclear if the sir1 phenotype could be fully restored by further inhibiting GCL activity and if the activation of glutathione synthesis in sir1 would exacerbate the growth phenotype.
Decreased glutathione synthesis caused by the cad2 mutation drives significant accumulation of Cys and enhances S flux into protein synthesis. However, Cys levels are not affected in the sir1 mutant, whereas O-acetyl-Ser and sulfate accumulate due to activation of sulfate uptake (Speiser et al., 2018). Using flux analysis after feeding [35S]-sulfate, Speiser et al. showed that de novo incorporation of sulfate is dramatically decreased in sir1, driving depletion of S assimilation into Cys. Under these limiting conditions, Cys is used more efficiently for protein synthesis than for glutathione synthesis (Fig. 1). When glutathione synthesis is inhibited, total S assimilation and incorporation into Cys are enhanced in sir1 cad2 relative to sir1. Thus, a significant repartitioning of S from glutathione synthesis into protein translation is detected, driving an increase of protein S in the sir1 cad2 mutants and alleviating growth inhibition by the sensor kinase Target of Rapamycin (TOR; Speiser et al., 2018). Nevertheless, it is unclear how inhibition of GCL activity activates TOR signaling.
The discovery of GCL function as a switch that enhances the flux of de novo assimilated sulfate into proteins is relevant in a context of development and signaling in optimal growth conditions. However, in a context of environmental stress, the scenario may be somewhat different. While S in soluble proteins is estimated to be 6 to 10 times greater than the glutathione pool in optimal conditions (Queval et al., 2009), the difference can be shifted in oxidative stress conditions. For instance, ozone exposure or enhanced intracellular H2O2 can drive high accumulation of glutathione (Bick et al., 2001; Mhamdi et al., 2010), associated with up-regulation of Cys synthesis in the chloroplasts (Queval et al., 2009). Hence, the balance between S in glutathione and in proteins may change in conditions that promote cellular oxidation, with flux favoring glutathione synthesis in stress conditions. It remains unclear how this regulatory mechanism might affect protein synthesis in plants strongly overexpressing GCL. Increased glutathione in some of these plants has been shown to be associated with unexpected increases in the cellular oxidation state and the induction of defense pathways (Creissen et al., 1999; Matern et al., 2015), effects that are as yet not fully understood.
Upstream of Cys, S can be channeled to 5′-phosphoadenosine 3′-phosphate, 5′-phosphoadenosine 3′-phosphosulfate, and secondary compounds such as glucosinolates in the Brassicaceae (Fig. 1). The higher demand of S in leaves during stress is probably required to support the synthesis of defense and signaling metabolites. Like glutathione, 5′-phosphoadenosine 3′-phosphate and glucosinolates play important roles in triggering appropriate responses to stress (Noctor et al., 2012; Chan et al., 2013; Bruggeman et al., 2016). Moreover, S assimilation and Cys synthesis are partitioned between subcellular compartments. Current models evoke a scenario where chloroplasts generate reduced sulfide, mitochondria deliver O-acetyl-Ser, and the cytosol produces most of the Cys (Takahashi et al., 2011). The study by Speiser et al. (2018) adds an additional layer of complexity to the regulation of S metabolism by intermediate metabolites of the assimilation pathway to ensure efficient use of S and to fine-tune retrograde signaling, growth, and stress responses.
REFERENCES
Bick JA, Setterdahl AT, Knaff DB, Chen Y, Pitcher LH, Zilinskas BA, Leustek T (2001) Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry 40: 9040–9048
Bruggeman Q, Mazubert C, Prunier F, Lugan R, Chan KX, Phua SY, Pogson BJ, Krieger-Liszkay A, Delarue M, Benhamed M, et al. (2016) Chloroplast activity and 3′phosphadenosine 5′phosphate signaling regulate programmed cell death in Arabidopsis. Plant Physiol 170: 1745–1756
Chan KX, Wirtz M, Phua SY, Estavillo GM, Pogson BJ (2013) Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci 18: 18–29
Cobbett CS, May MJ, Howden R, Rolls B (1998) The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in γ-glutamylcysteine synthetase. Plant J 16: 73–78
Creissen G, Firmin J, Fryer M, Kular B, Leyland N, Reynolds H, Pastori G, Wellburn F, Baker N, Wellburn A, et al. (1999) Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell 11: 1277–1292
Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174
Hopkins L, Parmar S, Błaszczyk A, Hesse H, Hoefgen R, Hawkesford MJ (2005) O-acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiol 138: 433–440
Khan MS, Haas FH, Samami AA, Gholami AM, Bauer A, Fellenberg K, Reichelt M, Hänsch R, Mendel RR, Meyer AJ, et al. (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22: 1216–1231
Matern S, Peskan-Berghoefer T, Gromes R, Kiesel RV, Rausch T (2015) Imposed glutathione-mediated redox switch modulates the tobacco wound-induced protein kinase and salicylic acid-induced protein kinase activation state and impacts on defence against Pseudomonas syringae. J Exp Bot 66: 1935–1950
Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis-Bourguet E, Renou JP, et al. (2010) Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol 153: 1144–1160
Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH (2012) Glutathione in plants: an integrated overview. Plant Cell Environ 35: 454–484
Queval G, Thominet D, Vanacker H, Miginiac-Maslow M, Gakière B, Noctor G (2009) H2O2-activated up-regulation of glutathione in Arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol Plant 2: 344–356
Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184