Escape from Centromere Land
As plant biologists, we do love to consider the physiological adaptations plants have made to being sessile organisms—unlike animals, plants cannot move away from adverse environmental conditions such as high temperature, etc. We commonly consider such responses for organisms, but what about genes? Over evolutionary time (much as environmental conditions change rapidly and unpredictably), the genomic landscape changes, with regions undergoing DNA-level rearrangements, epigenetic changes in DNA and histone modifications, and functional changes, such as movement of centromeres into adjacent areas, or estabilishment of neocentromeres in new areas (reviewed in Comai et al., 2017). How do these changes affect a gene’s environment and ability to function? And, do genes move along, or remain and adapt?
 One particularly interesting place to examine this question is the centromere, where the repetitive, often heterochromatic environment undergoes changes in primary sequence, epigenetic modification, and even function as centromere positions shift over time. Examining genome-level changes in centromere structure requires a high-confidence genomic assembly of these highly repetitive regions. Indeed the combination of large, variable tracts of tandem repeats and transposons has remained a major challenge, even for most “complete” genomes. To remedy this, Liao et al. (2018) produce a 12.4 Mb of sequence of Oryza brachyantha centromeres, pyrosequencing 126 bacterial artificial chromosome clones and validating the assembly by comparison to maps produced by optical mapping. Their assemblies extended into the tandem arrays of the O. brachyantha centromere satellite sequence CentO-F. They further used chromatin immunoprecipitation and sequencing, with antibodies against the centromere-specific histone CenH3 to identify the functional centromere regions in O. brachyantha. The high-confidence sequence and functional verification allowed them to compare the centromeres of O. brachyantha to those of cultivated rice, O. sativa (using a japonica and an indica cultivar). These two rice species diverged roughly 15 million years ago—a brief timeframe for protein-coding genes. However, synteny analysis, with related grass species (e.g. Leersia perrieri, Brachypodium distachyon, and Setaria italica) as outgroups, showed that although the general positions of the centromeres were conserved, the centromere landscape in these two species underwent substantial shifts, with seven pericentric inversions (>50 kb) in O. sativa and ten in O. brachyantha. These events included three inversions that caused the position of the centromere to shift, and at least one centromere-repositioning event where the centromere moved about 400 kb in O. brachyantha. These rapid changes would challenge any but the most tolerant, or nimble genomic element.
One particularly interesting place to examine this question is the centromere, where the repetitive, often heterochromatic environment undergoes changes in primary sequence, epigenetic modification, and even function as centromere positions shift over time. Examining genome-level changes in centromere structure requires a high-confidence genomic assembly of these highly repetitive regions. Indeed the combination of large, variable tracts of tandem repeats and transposons has remained a major challenge, even for most “complete” genomes. To remedy this, Liao et al. (2018) produce a 12.4 Mb of sequence of Oryza brachyantha centromeres, pyrosequencing 126 bacterial artificial chromosome clones and validating the assembly by comparison to maps produced by optical mapping. Their assemblies extended into the tandem arrays of the O. brachyantha centromere satellite sequence CentO-F. They further used chromatin immunoprecipitation and sequencing, with antibodies against the centromere-specific histone CenH3 to identify the functional centromere regions in O. brachyantha. The high-confidence sequence and functional verification allowed them to compare the centromeres of O. brachyantha to those of cultivated rice, O. sativa (using a japonica and an indica cultivar). These two rice species diverged roughly 15 million years ago—a brief timeframe for protein-coding genes. However, synteny analysis, with related grass species (e.g. Leersia perrieri, Brachypodium distachyon, and Setaria italica) as outgroups, showed that although the general positions of the centromeres were conserved, the centromere landscape in these two species underwent substantial shifts, with seven pericentric inversions (>50 kb) in O. sativa and ten in O. brachyantha. These events included three inversions that caused the position of the centromere to shift, and at least one centromere-repositioning event where the centromere moved about 400 kb in O. brachyantha. These rapid changes would challenge any but the most tolerant, or nimble genomic element.
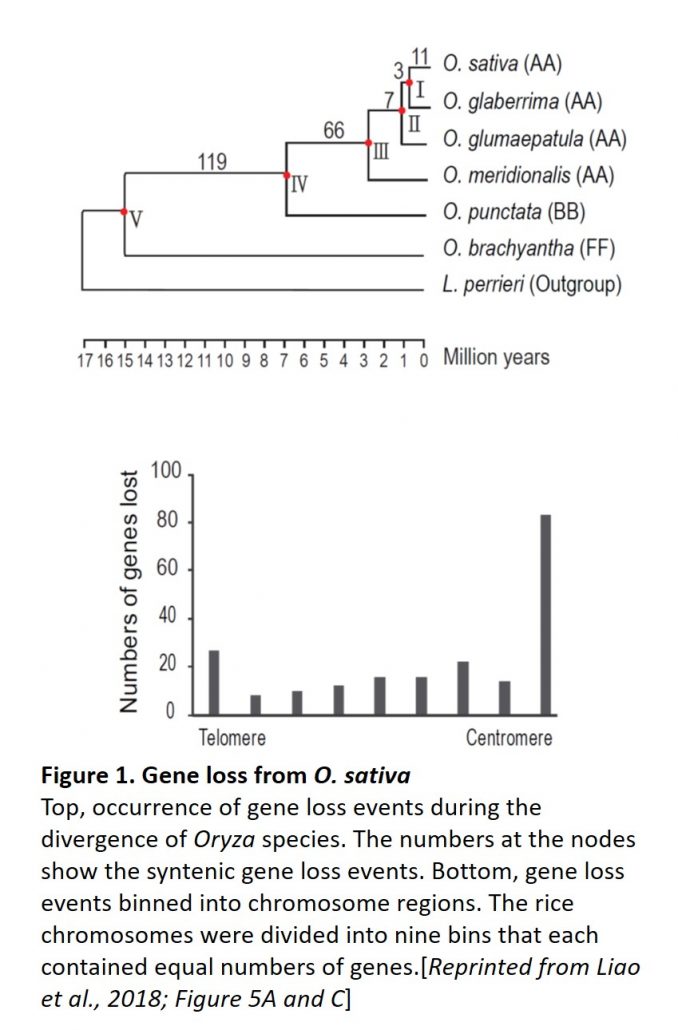
Are genomic conditions in the centromere unfavorable for protein-coding genes and do genes stay put like sessile plants, or move away like animals? Centromeres may be inhospitable environments for protein-coding genes because of their incompatibility with gene expression (although centromeres undergo pervasive transcription), as well as their frequent rearrangements and double-stranded breaks that can lead to position-effect variegation. To examine gene loss in O. sativa centromeres, the authors identified low-copy O. brachyantha genes with orthologs in L. perrieri and/or Brachypodium distachyon that were absent in the O. sativa centromeres. Comparison with other Oryza species allowed the authors to track the occurrence of these gene loss events (Figure). Binning the events into chromosome segments identified gene loss events scattered over the chromosomes, but enriched near the centromeres (Figure). Also, regions that underwent centromere movement lost more genes in the new location of the centromere. Moreover, the O. sativa genome contains new (i.e. recent) copies of many of the “lost” genes (many of which encode essential proteins) in other genomic locations, likely as a result of segmental duplications and loss of the centromere copy. Therefore, O. sativa genes were preferentially lost from centromere regions and added in non-centromere regions—hardly sessile at all! Moreover, the authors identified gene gain at O. sativa centromeres, but the genes added to centromeres appeared to have newly emerged, rather than being duplicated from pre-existed genes before the divergence O. brachyantha and O. sativa. These intriguing results show that selection drives many genes to escape from the centromere; identification of the mechanisms that allow some genes to remain in this environment remains an interesting question for future work.
REFERENCES
Comai, L., Maheshwari, S., and Marimuthu, M.P.A. (2017) Plant Centromeres. Current Opinion in Plant Biology 36: 158-167.
Liao, Y., Zhang, X., Li, B., Liu, T., Chen, J., Bai, Z., Wang, M., Shi, J., Walling, J.G., Wing, R.A., Jiang, J., Chen, M. (2018). Comparison of Oryza sativa and Oryza brachyantha genomes reveals selection-driven gene escape from the centromeric regions. Plant Cell DOI: https://doi.org/10.1105/tpc.18.00163.



