Enzyme helps as a transcription factor in lignin production
In poplars a protein appears to have a bizarre double function: it makes building blocks for amino acids but also regulates the production of lignin. It could be a new way to create lignin-poor poplars, according to a publication in The Plant Cell .
Such lignin-poor trees are an old idea that has been studied for at least ten years, especially in Flanders . Poplars grow quickly and produce large amounts of cellulose that you can ferment to bio-ethanol. But the lignin fraction, which gives the wood its strength, prevents the extraction of that cellulose. Wout Boerjan and colleagues from Ghent University have already managed to reduce that lignin fraction by blocking a few other enzymes, but there is still plenty of room for alternatives – especially as the Flemish enzymes have been patented.
 The alternative presented by Wellington Muchero, Meng Xie and colleagues from Oak Ridge National Laboratory is called 5-enolpyruvylshikimate 3-phosphate synthase, abbreviated as PtrEPSP. Ptr stands for Populus trichocarpa , the Latin name for the poplar. The name already betrays that it is part of the Shikimate synthesis route , which ultimately yields the aromatic amino acids phenylalanine, tyrosine and tryptophan.
The alternative presented by Wellington Muchero, Meng Xie and colleagues from Oak Ridge National Laboratory is called 5-enolpyruvylshikimate 3-phosphate synthase, abbreviated as PtrEPSP. Ptr stands for Populus trichocarpa , the Latin name for the poplar. The name already betrays that it is part of the Shikimate synthesis route , which ultimately yields the aromatic amino acids phenylalanine, tyrosine and tryptophan.
Phenylalanine is in turn the starting point for the phenylpropanoid route , which eventually yields lignin. There are very different enzymes involved. That the lignin production decreases if you block PtrEPSP is not illogical, but only because you create a shortage of phenylalanine.
Incidentally, the lignin production decreases slightly more drastically because the tree does not survive such an intervention, due to lack of amino acids as building blocks for its proteins. It is not for nothing that EPSP is the target of the famous herbicide glyphosate.
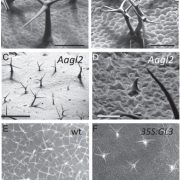
But with association mapping techniques , they have now demonstrated a second connection in Oak Ridge. It comes down to the fact that overproduction of PtrEPSP also yields more lignin, and also abnormal expression of some other genes that have to do with lignin production.
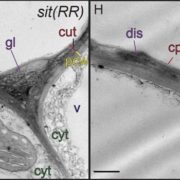
Further research showed that PtrEPSP accumulates in such cells in the nucleus, where it has nothing to look for on the Shikimineroute. It inhibits a gene called PtrhAT, and that cranks the entire phenylpropanoid pathway.
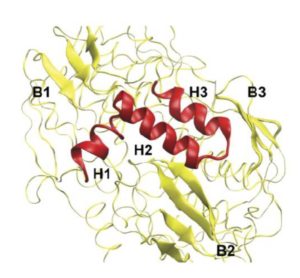
Then it turned out that poplars create two types of PtrEPSP. The genetic codes are very similar and the resulting proteins both function as 5-enolpyruvylshikimate 3-phosphate synthase. But one of the two codes is a lot longer and at the N-terminus of that protein, called PtrEPSP-TF, hangs an extra piece of 128 amino acids. And that end has the specific form (an ‘HTH motif’) that causes the enzyme to enter the cell nucleus and to bind to the DNA there in order to inhibit the expression of PtrhAT.
In retrospect, PtrEPSP-TF appears to be mainly expressed in places where the tree requires extra lignin. The short version, PtrEPSP-SY, can be seen in areas where amino acids are produced, ie near chloroplasts that provide the necessary input.
As an enolpyruvylshikimate 3-phosphate synthase, PtrEPSP-TF performs a lot less than PtrEPSP-SY, and it is suspected that this function is a relic of the past. Once the complete genome of poplars has inadvertently been doubled, and apparently the evolution has subsequently reused one of the two sets of EPSP genes for a completely new task.
source: ORNL