Deep learning for multi-task plant phenotyping
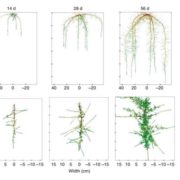
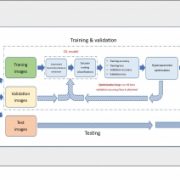
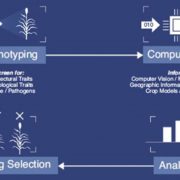
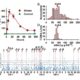
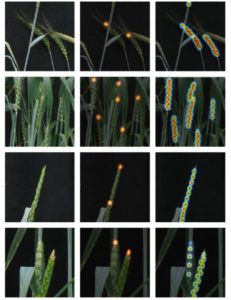 Despite the significant developments in computational biology and modern plant breeding, crop phenotyping poses challenges for automation. Previous machine learning approaches of plant phenotyping have mainly focused on leaf counting in rosette forms or leaf segmentation, and rely on large datasets not particularly useful for monocot crops. Pound et al. have recently described a valuable dataset tool called ACID (The Annotated Crop Image Dataset) that is primarily developed on wheat to localize spikes and spikelets. The dataset is based on a representation of 520 images of wheat plants with a wide range of plant and spike phenotypes. Each plant has been annotated individually to determine the location of spike and individual spikelets. The network performs simultaneous classification of images, with a detailed annotation of spike and spikelets and classification of awned plants. This is an interesting advancement in machine learning phenotyping (http://plantimages.nottingham.ac.uk/) and will be useful for scientists in the improvement of crop phenotyping on all plants with architecture similar to wheat. With the advancement of wheat breeding with varied phenotypic lines, the deep learning machine approach will be necessary for accurate phenotypic quantification with a view to predict yield. (Summary by Amey Redkar) bioRxiv 10.1101/204552
Despite the significant developments in computational biology and modern plant breeding, crop phenotyping poses challenges for automation. Previous machine learning approaches of plant phenotyping have mainly focused on leaf counting in rosette forms or leaf segmentation, and rely on large datasets not particularly useful for monocot crops. Pound et al. have recently described a valuable dataset tool called ACID (The Annotated Crop Image Dataset) that is primarily developed on wheat to localize spikes and spikelets. The dataset is based on a representation of 520 images of wheat plants with a wide range of plant and spike phenotypes. Each plant has been annotated individually to determine the location of spike and individual spikelets. The network performs simultaneous classification of images, with a detailed annotation of spike and spikelets and classification of awned plants. This is an interesting advancement in machine learning phenotyping (http://plantimages.nottingham.ac.uk/) and will be useful for scientists in the improvement of crop phenotyping on all plants with architecture similar to wheat. With the advancement of wheat breeding with varied phenotypic lines, the deep learning machine approach will be necessary for accurate phenotypic quantification with a view to predict yield. (Summary by Amey Redkar) bioRxiv 10.1101/204552