A Method to the Madness: Using Persistent Homology to Measure Plant Morphology
As the study of plant variation and characterization moves from measurements of largely organismal-level polymorphisms to those of cellular- and molecular-level traits, the need for morphological measuring tools that incorporate complex trait information is clear. While quantitative methodologies have improved, current methods rely on the researcher evaluating what morphological metrics to measure, and this results in a somewhat biased collection of values for univariate or multivariate analysis (Topp et al., 2013). The models that rely on the compilation of standard univariate measurements are not flexible enough to accommodate a large amount of variation in plant traits, which limits our understanding of genetic and environmental contributions to plant morphology (Chitwood and Topp, 2015).
In this issue, Li et al. (2018) address the limitations in morphological evaluation with an innovative mathematical framework using a tool from the field of topological data analysis called persistent homology (PH) that can more accurately quantify phenotypic variation than other quantitative methods. In PH, topological features are monitored at different spatial resolutions to allow for traits or characters to be identified during analysis rather than prior to measurement. PH measurements can be designed to be orientation-independent and resistant to the noise that often accompanies data collection from two-dimensional (2D) images. In this way, PH captures multiple and comprehensive measurements better than uni- or multivariate systems because it constructs a single metric value from several morphological features. The PH metrics therefore capture more information about plant morphology in a comprehensive way than a single trait, and are not reliant on orientation or size, which is a significant factor in the ability of PH to integrate multiple morphological traits into a single descriptor that can be applied to different tissue types, growth conditions, and plant species.
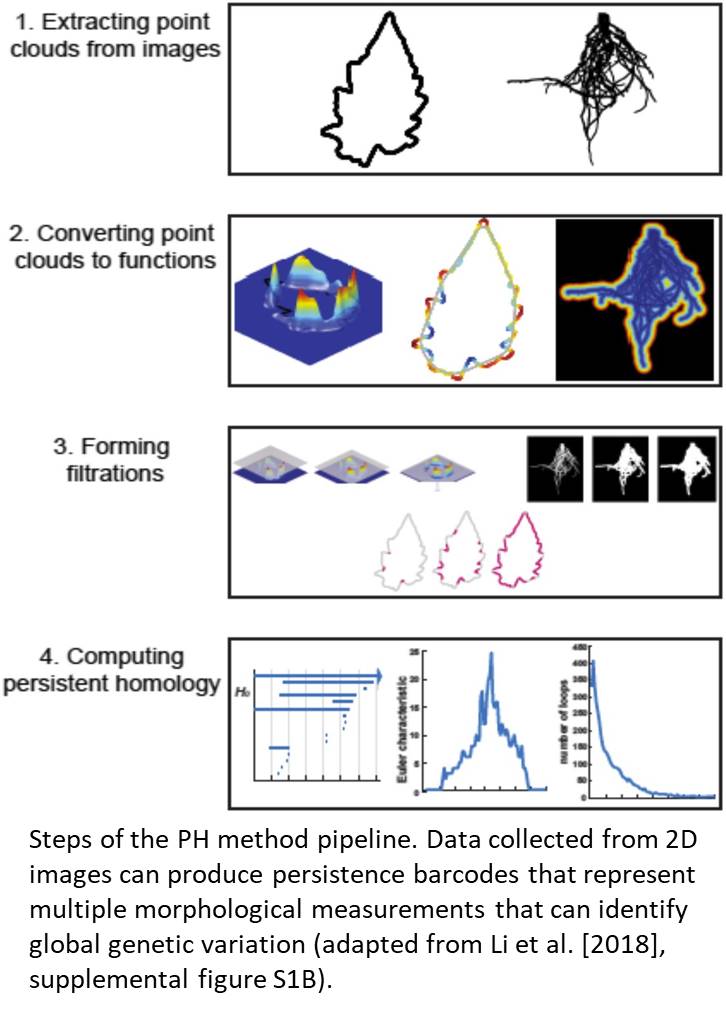 The authors tested how PH could reveal pleiotropic genetic components of natural variation in shoot and root morphologies by measuring aspects of leaf shape, leaflet serration, and root architecture in a wild tomato (Solanum pennellii) introgression population. Using the PH method on leaf shape normalized leaf contours and removed noise, which are time-consuming steps inherent to traditional 2D image analysis methods. Developing a PH function specific to variation in leaflet serrations helped to more specifically define variations as a local aspect of leaf shape and contour. Root architecture was assessed in harvested root systems of field-grown plants; PH utilized branching and branch crossing patterns from 2D images as a form of topological homology that informed the resulting feature value. After these topological features were analyzed, persistence barcodes that capture multiple morphological variations in a single metric were used to detect global genetic changes in shoot and root organization (Fig. 1). Similar effect sizes for genotypic discrimination of complex traits were observed between the PH and multivariate approaches; however, PH did a better job at describing large-effect heritable traits for the complex shapes evaluated. Moreover, PH identified more unique quantitative trait loci than did traditional methods. Because this analysis used root and shoot trait variation, the PH method also identified overlapping quantitative trait loci that were associated with both leaf shape and root architecture values, indicating that this method can identify genetic attributes in one part of the plant that affect other parts as well.
The authors tested how PH could reveal pleiotropic genetic components of natural variation in shoot and root morphologies by measuring aspects of leaf shape, leaflet serration, and root architecture in a wild tomato (Solanum pennellii) introgression population. Using the PH method on leaf shape normalized leaf contours and removed noise, which are time-consuming steps inherent to traditional 2D image analysis methods. Developing a PH function specific to variation in leaflet serrations helped to more specifically define variations as a local aspect of leaf shape and contour. Root architecture was assessed in harvested root systems of field-grown plants; PH utilized branching and branch crossing patterns from 2D images as a form of topological homology that informed the resulting feature value. After these topological features were analyzed, persistence barcodes that capture multiple morphological variations in a single metric were used to detect global genetic changes in shoot and root organization (Fig. 1). Similar effect sizes for genotypic discrimination of complex traits were observed between the PH and multivariate approaches; however, PH did a better job at describing large-effect heritable traits for the complex shapes evaluated. Moreover, PH identified more unique quantitative trait loci than did traditional methods. Because this analysis used root and shoot trait variation, the PH method also identified overlapping quantitative trait loci that were associated with both leaf shape and root architecture values, indicating that this method can identify genetic attributes in one part of the plant that affect other parts as well.
Compared with conventional approaches, PH is a flexible, high-throughput method of evaluating variation in plant morphology that is open to automation and improves upon current uni- and multivariate methods. Perhaps most importantly, PH could be used to measure dynamic morphologies that change over time and thereby further our understanding of how plant features are controlled by both genetic and environmental factors.
REFERENCES
Chitwood DH, Topp CN (2015) Revealing plant cryptotypes: defining meaningful phenotypes among infinite traits. Curr Opin Plant Biol 24: 54–60
Li M, Frank MH, Coneva V, Mio W, Chitwood DH, Topp CN (2018) The persistent homology mathematical framework provides enhanced genotype-to-phenotype associations for plant morphology. Plant Physiol 177: 1382-1395.
Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee C-R, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110: E1695–E1704