Plant Science Research Weekly: December 13
This week we start with a short summary contributed by Charlotte Gommers hghlighting several papers published in 2019 that revealed new insights into an enigmatic plant gene, GUN1.
Research update: GUN1 hit the mark in 2019
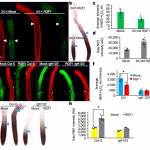
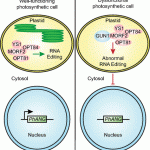
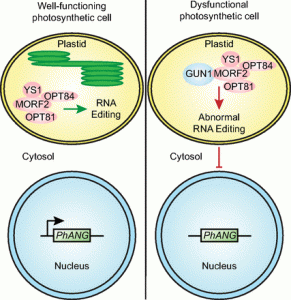 Chloroplasts are able to sense and respond to environmental signals. They send retrograde signals that inform the nucleus about their developmental stage and integrity. In response, the nucleus adjusts gene expression to optimize chloroplast recovery and adaptation. Back in 1993, a screen for genotypes that lack the repression of nuclear photosynthesis genes in response to chemical inhibition of chloroplasts revealed the Arabidopsis thaliana genomes uncoupled (gun) mutants (Susek et al., 1993). In the following years, characterization of the targeted proteins revealed that GUN2 through GUN6 act in the tetrapyrrole biosynthesis pathway, but GUN1 was the exception. This pentatricopeptide repeat protein appeared essential during the early phases of chloroplast development and biogenic retrograde signaling, but its exact function remained speculative. During the Jacques Monod conference on Retrograde Signalling from Endosymbiotic Organelles in 2018, someone even posed that maybe “GUN1 is actually more important to us, scientists, than to plants”.
Chloroplasts are able to sense and respond to environmental signals. They send retrograde signals that inform the nucleus about their developmental stage and integrity. In response, the nucleus adjusts gene expression to optimize chloroplast recovery and adaptation. Back in 1993, a screen for genotypes that lack the repression of nuclear photosynthesis genes in response to chemical inhibition of chloroplasts revealed the Arabidopsis thaliana genomes uncoupled (gun) mutants (Susek et al., 1993). In the following years, characterization of the targeted proteins revealed that GUN2 through GUN6 act in the tetrapyrrole biosynthesis pathway, but GUN1 was the exception. This pentatricopeptide repeat protein appeared essential during the early phases of chloroplast development and biogenic retrograde signaling, but its exact function remained speculative. During the Jacques Monod conference on Retrograde Signalling from Endosymbiotic Organelles in 2018, someone even posed that maybe “GUN1 is actually more important to us, scientists, than to plants”.
But 2019 has been an enlightening year for GUN1 as we finally learned loads about its function. It turns out, GUN1 pays a very important role in many different processes in the chloroplast. It 1) interacts with MULTIPLE ORGANELLAR RNA EDITING FACTOR2 (MORF2) and is involved in plastid RNA editing (Zhao et al., 2019), 2) regulates protein import into the chloroplast via its interaction with the chloroplast HEAT SHOCK PROTEIN70-1 (cpHSC70-1) (Wu et al., 2019), 3) maintains plastid protein homeostasis (Marino et al., 2019), 4) is involved in NUCLEAR ENCODED POLYMERASE (NEP) mediated transcription in the chloroplast (Tadini et al., 2019) and finally, 5) directly affects the flux through the tetrapyrrole biosynthesis pathway by directly interacting with heme and other porphyrins, suggesting that gun1 might be not as different from gun2 through gun6 after all (Shimizu et al., 2019). The work published on GUN1 function the past year is a breakthrough for retrograde signaling research. I look forward to finding out what more the future beholds for GUN1! (Contributed by Charlotte Gommers).
Citations: Marino G, Naranjo B, Wang J, Penzler JF, Kleine T, Leister D (2019) Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J 99: 521-535; Shimizu T, Kacprzak SM, Mochizuki N, Nagatani A, Watanabe S, Shimada T, Tanaka K, Hayashi Y, Arai M, Leister D, Okamoto H, Terry MJ, Masuda T (2019) The retrograde signaling protein GUN1 regulates tetrapyrrole biosynthesis. Proc. Natl. Acad. Sci. USA: 201911251; Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787-799; Tadini L, Peracchio C, Trotta A, Colombo M, Mancini I, Jeran N, Costa A, Faoro F, Marsoni M, Vannini C, Aro E-M, Pesaresi P (2019) GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J. doi:10.1111/tpj.14585; Wu G-Z, Meyer EH, Richter AS, Schuster M, Ling Q, Schöttler MA, Walther D, Zoschke R, Grimm B, Jarvis RP, Bock R (2019) Control of retrograde signalling by protein import and cytosolic folding stress. Nature Plants 5: 525-538; Zhao X, Huang J, Chory J (2019) GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. USA: 116: 10162-10167
Review. Brassinosteroids: Multi-dimensional regulators of plant growth, development, and stress responses
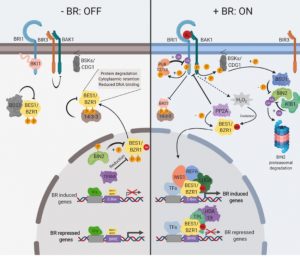 It’s been fifty years since the existence of brassinosteroids was shown, and about thirty since the awesome power of Arabidopsis genetics started to reveal the genes involved in its synthesis, perception and signaling. Nolan et al. review the highlights of brassinosteroid research and also more recent findings, particularly new insights into how it intersects with other hormones and signals, and its contributions to stress responses. The authors discuss how manipulations of brassinosteroid biology can be applied in crops, and identify key unresolved questions. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00335
It’s been fifty years since the existence of brassinosteroids was shown, and about thirty since the awesome power of Arabidopsis genetics started to reveal the genes involved in its synthesis, perception and signaling. Nolan et al. review the highlights of brassinosteroid research and also more recent findings, particularly new insights into how it intersects with other hormones and signals, and its contributions to stress responses. The authors discuss how manipulations of brassinosteroid biology can be applied in crops, and identify key unresolved questions. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00335
Perspective: Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens
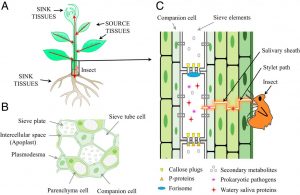 Much of our understanding of plant defense response is built upon the responses that occur in leaves. However, many pathogens colonize the phloem system, which is both nutrient-rich and provides an easy conduit for spreading systemically through the plant body. These phloem-inhabiting prokaryotic pathogens are introduced and spread through phloem-feeding insects. Jiang et al. describe the challenges in studying these pathogens, which can cause serious diseases including citrus greening disease (aka “huanglongbin” or HLB). Phloem-resident prokaryotic pathogens include phytoplasmas and spiroplasmas, which are notable both for having a reduced genome and being difficult (or impossible) to culture in vitro, contributing to the challenges of studying them. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1915396116. (This PNAS perspective is contributed by Sheng Yang He, who is also featured in a QnA https://doi.org/10.1073/pnas.1918514116).
Much of our understanding of plant defense response is built upon the responses that occur in leaves. However, many pathogens colonize the phloem system, which is both nutrient-rich and provides an easy conduit for spreading systemically through the plant body. These phloem-inhabiting prokaryotic pathogens are introduced and spread through phloem-feeding insects. Jiang et al. describe the challenges in studying these pathogens, which can cause serious diseases including citrus greening disease (aka “huanglongbin” or HLB). Phloem-resident prokaryotic pathogens include phytoplasmas and spiroplasmas, which are notable both for having a reduced genome and being difficult (or impossible) to culture in vitro, contributing to the challenges of studying them. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1915396116. (This PNAS perspective is contributed by Sheng Yang He, who is also featured in a QnA https://doi.org/10.1073/pnas.1918514116).
Unraveling cis and trans regulatory evolution during cotton domestication
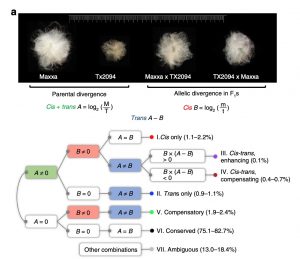 Polyploidization leads to a myriad of changes in gene expression and organization of genomes and can supply the material for speciation, adaptation, and morphological innovation. The most cultivated cotton species, Gossypium hirsutum, is an allotetraploid species (AD genome) containing two subgenomes (A from African and D from American diploid progenitors). Bao et al. have analyzed 27 RNA-seq libraries of four G. hirsutum accessions (wild and domesticated parental cotton and their reciprocal F1 hybrids) in two fiber developmental stages (fibers are single-celled seed trichomes). Through this they identified potential cis- and trans-regulatory element variation during cotton domestication, their inheritance patterns, and their association in co-expression and regulatory networks that are imperative for fiber development. The findings described by the study enlighten the evolutionary story of cotton fiber, and they will be very useful for agricultural improvement strategies in G. hirsutum and other polyploid species. (Summary by Sarah M. Nardeli) Nature Comms. 10.1038/s41467-019-13386-w
Polyploidization leads to a myriad of changes in gene expression and organization of genomes and can supply the material for speciation, adaptation, and morphological innovation. The most cultivated cotton species, Gossypium hirsutum, is an allotetraploid species (AD genome) containing two subgenomes (A from African and D from American diploid progenitors). Bao et al. have analyzed 27 RNA-seq libraries of four G. hirsutum accessions (wild and domesticated parental cotton and their reciprocal F1 hybrids) in two fiber developmental stages (fibers are single-celled seed trichomes). Through this they identified potential cis- and trans-regulatory element variation during cotton domestication, their inheritance patterns, and their association in co-expression and regulatory networks that are imperative for fiber development. The findings described by the study enlighten the evolutionary story of cotton fiber, and they will be very useful for agricultural improvement strategies in G. hirsutum and other polyploid species. (Summary by Sarah M. Nardeli) Nature Comms. 10.1038/s41467-019-13386-w
NONSTOP GLUMES 1 regulates spikelet development in rice
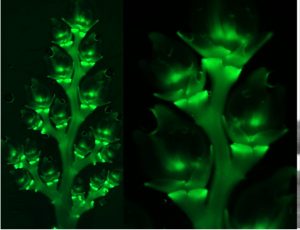 Recently, several genes affecting rice architecture have been identified that may increase yields by increasing the number of grains formed, but as yet the genetic control of rice inflorescence architecture and organ identity is still being worked out. Zhuang et al. identified mutants of the NONSTOP GLUMES 1 (NSG1) gene (encoding a transcription factor) that affect spikelet development and shed light on the gene regulatory networks that specify floral and non-floral organs. Loss-of-function nsg1 mutants show abnormalities and partial conversion of the outer spikelet organs (glumes, lemmas, palea). Additional analysis showed ectopic expression of several organ identity genes in the mutant, including LEAFY HULL STERILE 1 (LHS1), DROOPING LEAF (DL), and MOSAIC FLORAL ORGANS 1 (MFO1). A greater understanding of the genetic and molecular mechanism of spikelet development could provide additional tools for increasing grain numbers and yields. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00682
Recently, several genes affecting rice architecture have been identified that may increase yields by increasing the number of grains formed, but as yet the genetic control of rice inflorescence architecture and organ identity is still being worked out. Zhuang et al. identified mutants of the NONSTOP GLUMES 1 (NSG1) gene (encoding a transcription factor) that affect spikelet development and shed light on the gene regulatory networks that specify floral and non-floral organs. Loss-of-function nsg1 mutants show abnormalities and partial conversion of the outer spikelet organs (glumes, lemmas, palea). Additional analysis showed ectopic expression of several organ identity genes in the mutant, including LEAFY HULL STERILE 1 (LHS1), DROOPING LEAF (DL), and MOSAIC FLORAL ORGANS 1 (MFO1). A greater understanding of the genetic and molecular mechanism of spikelet development could provide additional tools for increasing grain numbers and yields. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00682
Autoregulation of RCO by low-affinity binding modulates cytokinin action and shapes leaf diversity
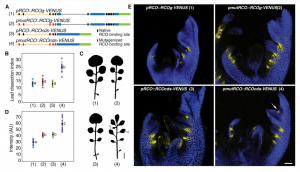 REDUCED COMPLEXITY (RCO) is the main homeodomain transcription factor (TF) responsible for leaf complexity in cruciferous plants. RCO is lost in the model Arabidopsis with simple leaves but present in the genome of Cardamine hirsuta, a good model for leaf morphology traits. However, little is known about RCO’s genome-wide targets. In this paper, Hajheidari et al. address this question using ChiP-seq and RNA-seq in C. hirsuta. Among the identified targets, the authors found that RCO regulates cytokinin signaling, activating the expression of genes involved in CK homeostasis and signaling and locally increasing CK activity in the leaflets. Additionally, RCO expression is autoregulated by repressing itself. This observation is consistent with gene expression analysis and it is crucial to the precise expression of the gene in the leaflets. Instead of a single DNA binding site in the promoter region, the authors found 11 non-canonical low-affinity binding motifs along a distal enhancer and intronic region that mediating the negative feedback-loop and contribute to the fine-tuning of the gene expression. Low-affinity binding sites put in question the simplified logic of transcriptional regulation and can coexist with other regulatory modes in the context of different loci. This kind of interaction was also described for other homeodomain TFs in animals. (Summary by Facundo Romani) Curr. Biol. 10.1016/j.cub.2019.10.040
REDUCED COMPLEXITY (RCO) is the main homeodomain transcription factor (TF) responsible for leaf complexity in cruciferous plants. RCO is lost in the model Arabidopsis with simple leaves but present in the genome of Cardamine hirsuta, a good model for leaf morphology traits. However, little is known about RCO’s genome-wide targets. In this paper, Hajheidari et al. address this question using ChiP-seq and RNA-seq in C. hirsuta. Among the identified targets, the authors found that RCO regulates cytokinin signaling, activating the expression of genes involved in CK homeostasis and signaling and locally increasing CK activity in the leaflets. Additionally, RCO expression is autoregulated by repressing itself. This observation is consistent with gene expression analysis and it is crucial to the precise expression of the gene in the leaflets. Instead of a single DNA binding site in the promoter region, the authors found 11 non-canonical low-affinity binding motifs along a distal enhancer and intronic region that mediating the negative feedback-loop and contribute to the fine-tuning of the gene expression. Low-affinity binding sites put in question the simplified logic of transcriptional regulation and can coexist with other regulatory modes in the context of different loci. This kind of interaction was also described for other homeodomain TFs in animals. (Summary by Facundo Romani) Curr. Biol. 10.1016/j.cub.2019.10.040
CSN5 of COP9 signalosome modulates heat-response of Arabidopsis
 COP9 signalosome (CSN) is a conserved multi-subunit complex in higher eukaryotes that was originally identified as a regulator of plant photomorphogenesis. It functions by regulating ubiquitin-mediated protein stability, through the deneddylase enzymatic acitivity of the CSN5 subunit. CSN5 is encoded by two genes, and the csn5a-1 mutant shows a deficiency of deneddylation, leading to hyper-neddylation of the ubiquitin ligase CUL1 (a ubiquitin scaffold protein). Recently, Singh et al. found that heat-treated csn5a-1 mutants showed an increase in cell size, ploidy, photosynthetic activity, and an upregulation of genes connected to the auxin response. Because auxin signaling is enhanced in heat-treated Arabidopsis seedlings, this shows that CSN5 contributes to maintaining auxin homeostasis in response to heat. This study opens new dimensions of CSN biology, as it explores the responses of COP9 signalosome in temperature responses. (Summary by Nanxun Qin) biomolecules 10.3390/biom9120805
COP9 signalosome (CSN) is a conserved multi-subunit complex in higher eukaryotes that was originally identified as a regulator of plant photomorphogenesis. It functions by regulating ubiquitin-mediated protein stability, through the deneddylase enzymatic acitivity of the CSN5 subunit. CSN5 is encoded by two genes, and the csn5a-1 mutant shows a deficiency of deneddylation, leading to hyper-neddylation of the ubiquitin ligase CUL1 (a ubiquitin scaffold protein). Recently, Singh et al. found that heat-treated csn5a-1 mutants showed an increase in cell size, ploidy, photosynthetic activity, and an upregulation of genes connected to the auxin response. Because auxin signaling is enhanced in heat-treated Arabidopsis seedlings, this shows that CSN5 contributes to maintaining auxin homeostasis in response to heat. This study opens new dimensions of CSN biology, as it explores the responses of COP9 signalosome in temperature responses. (Summary by Nanxun Qin) biomolecules 10.3390/biom9120805
Switching the direction of stem gravitropism by altering two amino acids in AtLAZY1
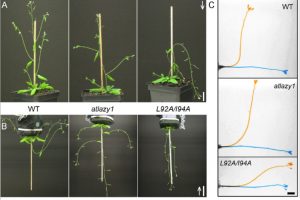 In a fascinating twist to the gravitropism story, Yoshihara and Spalding have managed to make a plant with shoots that don’t ignore gravity, but actually respond in the entirely inappropriate away. The LAZY genes were first identified from a rice mutant with a decreased response to gravity. In Arabidopsis there are five genes in this family, and most share several conserved sequences of unknown function. Using atlazy1 mutants, the authors tested proteins with mutations in each of the conserved region and found one that showed a flipped gravitropism, with shoots that selectively grow down (positive gravitropism) rather than up (negative gravitropism). Interestingly, the mutant plants show a normal response to asymmetric auxin, but in the mutant the auxin gradient is flipped relative to in wild-type plants. As yet the connection between AtLAZY1 and the auxin gradient is still being worked out. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01144
In a fascinating twist to the gravitropism story, Yoshihara and Spalding have managed to make a plant with shoots that don’t ignore gravity, but actually respond in the entirely inappropriate away. The LAZY genes were first identified from a rice mutant with a decreased response to gravity. In Arabidopsis there are five genes in this family, and most share several conserved sequences of unknown function. Using atlazy1 mutants, the authors tested proteins with mutations in each of the conserved region and found one that showed a flipped gravitropism, with shoots that selectively grow down (positive gravitropism) rather than up (negative gravitropism). Interestingly, the mutant plants show a normal response to asymmetric auxin, but in the mutant the auxin gradient is flipped relative to in wild-type plants. As yet the connection between AtLAZY1 and the auxin gradient is still being worked out. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.01144
RGF1 controls root meristem size through ROS signaling
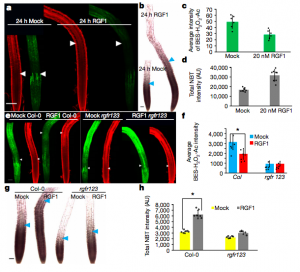 Stem cells are maintained in the stem-cell niche by intercellular interactions and signaling networks in which a peptide hormone, the root meristem growth factor 1 (RGF1), is involved. RGF1 is also important in the control of the size of the meristematic zone and in the stability of the PLETHORA (PLT) 1/2 proteins, which are required for stem cell maintenance; however, the downstream signaling mechanism are not well studied. Here, Yamada et al. demonstrate that in the developmental zones of the Arabidopsis root, the RGF1-receptor pathway controls the distribution of reactive oxygen species (ROS). In order to describe downstream factors in the RGF1 and ROS signalling pathway, the RGF1 transcriptome data was combined with developmental-zone-specific transcriptome data, highlighting the gene RGF1-INDUCIBLE TRANSCRIPTION FACTOR 1 (RITF1), predominantly expressed in the meristematic zone. The authors showed that RITF1 can modulate ROS signalling and root meristem size downstream of the RGF1 pathway. (Summarized by Francesca Resentini) Nature 10.1038/s41586-019-1819-6
Stem cells are maintained in the stem-cell niche by intercellular interactions and signaling networks in which a peptide hormone, the root meristem growth factor 1 (RGF1), is involved. RGF1 is also important in the control of the size of the meristematic zone and in the stability of the PLETHORA (PLT) 1/2 proteins, which are required for stem cell maintenance; however, the downstream signaling mechanism are not well studied. Here, Yamada et al. demonstrate that in the developmental zones of the Arabidopsis root, the RGF1-receptor pathway controls the distribution of reactive oxygen species (ROS). In order to describe downstream factors in the RGF1 and ROS signalling pathway, the RGF1 transcriptome data was combined with developmental-zone-specific transcriptome data, highlighting the gene RGF1-INDUCIBLE TRANSCRIPTION FACTOR 1 (RITF1), predominantly expressed in the meristematic zone. The authors showed that RITF1 can modulate ROS signalling and root meristem size downstream of the RGF1 pathway. (Summarized by Francesca Resentini) Nature 10.1038/s41586-019-1819-6
Increasing risks of multiple breadbasket failure under 1.5 and 2 °C global warming
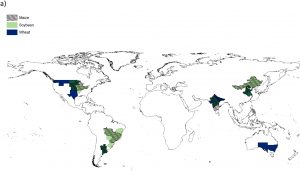 Crop yields are vulnerable to rising temperatures and changes in precipitation. Here, Gaupp et al. model the projected crop yields at 1.5 versus 2.0 degrees of additional warming, as part of the HAPPI experiment (perhaps a misnomer: Half a degree Additional warming, Prognosis and Projected Impacts). The authors look at risk of extreme weather events during the period when crops (wheat, soybean and maize) are growing and being harvested, and how these events will affect yield. They model local effects in each of the world’s major breadbasket regions (e.g., southwest Australia, Argentina, western China etc.) under current, 1.5 and 2 degree warming. Not surprisingly, both warming scenarios indicate yield losses, but what is particularly notable is that in the 2 degree warming scenario the impacts are likely to be felt across all regions, leading to global shortages. (Summary by Mary Williams) Agricultural Systems 10.1016/j.agsy.2019.05.010. On a similar note, here’s a new paper by Kornhuber et al that shows how Amplified Rossby waves enhance risk of concurrent heatwaves in major breadbasket regions (basically, circumglobal oscillations of the jet stream can lead to simultaneous heatwaves in several key food production areas at the same time), from Nature Climate Change 10.1038/s41558-019-0637-z.
Crop yields are vulnerable to rising temperatures and changes in precipitation. Here, Gaupp et al. model the projected crop yields at 1.5 versus 2.0 degrees of additional warming, as part of the HAPPI experiment (perhaps a misnomer: Half a degree Additional warming, Prognosis and Projected Impacts). The authors look at risk of extreme weather events during the period when crops (wheat, soybean and maize) are growing and being harvested, and how these events will affect yield. They model local effects in each of the world’s major breadbasket regions (e.g., southwest Australia, Argentina, western China etc.) under current, 1.5 and 2 degree warming. Not surprisingly, both warming scenarios indicate yield losses, but what is particularly notable is that in the 2 degree warming scenario the impacts are likely to be felt across all regions, leading to global shortages. (Summary by Mary Williams) Agricultural Systems 10.1016/j.agsy.2019.05.010. On a similar note, here’s a new paper by Kornhuber et al that shows how Amplified Rossby waves enhance risk of concurrent heatwaves in major breadbasket regions (basically, circumglobal oscillations of the jet stream can lead to simultaneous heatwaves in several key food production areas at the same time), from Nature Climate Change 10.1038/s41558-019-0637-z.