The Long Non-coding RNA ELENA1 Functions in Plant Immunity
Once seen as potential sequencing artifacts, long, non-coding RNAs (lncRNAs, >200 nucleotides) have gained recognition as important regulatory factors. LncRNAs are transcribed from a variety of genomic locations (introns, intergenic spaces, and coding regions) from the sense or antisense strand (reviewed in Chekanova, 2015). Moreover, lncRNAs function in cis or in trans and affect gene regulation transcriptionally or post-transcriptionally by diverse mechanisms, including recruiting factors that activate transcription or modify chromatin, serving as precursors of small RNAs, and even potentially affecting nuclear architecture. Plant lncRNAs function in RNA-directed DNA methylation (via production of small interfering RNAs), and developmental processes from phosphate signaling (via action as a target mimic to sequester a microRNA) to flowering time (via epigenetic regulation of FLOWERING LOCUS C expression).
In a microarray study, Liu et al. (2012) identified Arabidopsis thaliana polyadenylated lncRNAs induced by translation elongation factor Tu (elf18), a pathogen-associated factor that induces defense responses. Following up on this, Seo et al. (2017) characterized ELF18-INDUCED LONG-NONCODING RNA 1 (ELENA1). ELENA1, a 589-nucleotide lncRNA, is transcribed from an intergenic region. ELENA1 levels increased in response to elf18 and flagellin (flg22), but not in response to the defense hormone salicylic acid. Mutants affecting the receptors for elf18 and flg22 did not show an increase in ELENA1 levels in response to these inducers.
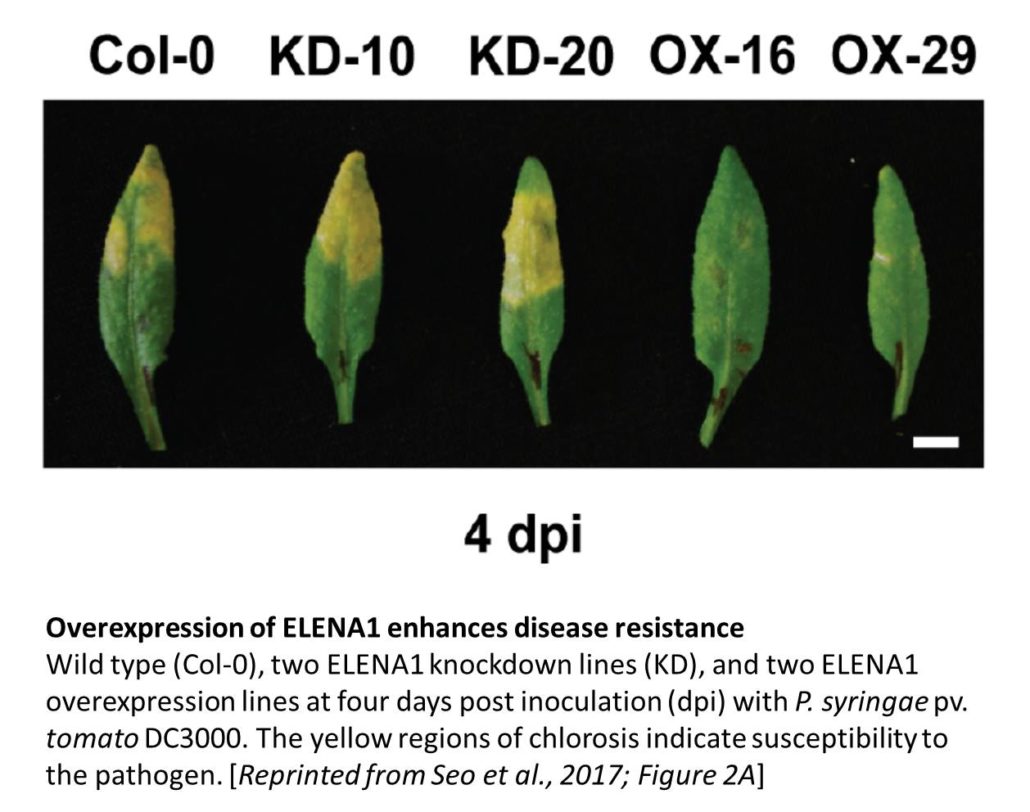 To examine ELENA1 function, the authors used an artificial microRNA to produce plants with reduced ELENA1 levels; these plants showed increased susceptibility to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (figure) and decreased expression of PATHOGENESIS-RELATED GENE 1 (PR1) during infection. By contrast, ELENA1 over-expression plants showed the opposite phenotypes. RNA-sequencing analysis and comparison of ELENA1 overexpressing plants with wild type, and uninduced with plants treated with elf18 showed that ELENA1 overexpression increased the expression of a subset of the genes induced in response to elf18. These included PR1 and PR2, genes encoding beta-1,3-glucanases affecting callose deposition, some salicylic acid-induced genes, and other genes involved in defense responses.
To examine ELENA1 function, the authors used an artificial microRNA to produce plants with reduced ELENA1 levels; these plants showed increased susceptibility to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (figure) and decreased expression of PATHOGENESIS-RELATED GENE 1 (PR1) during infection. By contrast, ELENA1 over-expression plants showed the opposite phenotypes. RNA-sequencing analysis and comparison of ELENA1 overexpressing plants with wild type, and uninduced with plants treated with elf18 showed that ELENA1 overexpression increased the expression of a subset of the genes induced in response to elf18. These included PR1 and PR2, genes encoding beta-1,3-glucanases affecting callose deposition, some salicylic acid-induced genes, and other genes involved in defense responses.
The ELENA1 RNA contains several small open reading frames, but mutagenesis of the start codons of these open reading frames did not change ELENA1 function: plants overexpressing the mutant version still showed strong induction of PR1 expression. This indicates that ELENA1 acts as RNA, not by encoding small peptides. Some lncRNAs activate transcription of their targets by interacting with Mediator, a complex of varying composition that mediates between transcription factors and RNA Polymerase II. Indeed, binding assays showed that ELENA1 interacts with several Mediator subunits, including MED19a. Also, med19a mutants have a phenotype similar to ELENA1 knockdown plants (decreased induction of PR1 in response to elf18 treatment). Moreover, RNA immunoprecipitation assays showed that the association of ELENA1 and MED19a increased in cells treated with elf18. Combinations of overexpression and knockdown/mutant lines showed that ELENA1 and MED19a function interdependently in inducing PR1 expression. Finally, chromatin immunoprecipitation assays showed that MED19a is enriched in a specific region of the PR1 promoter; this enrichment increases in ELENA1 overexpression plants and decreases in ELENA1 knockdown plants. Unlike many lncRNAs that act in cis to affect the transcriptional regulation of genes adjacent to the lncRNA-expressing locus, ELENA1 affects one nearby locus, but also acts in trans, affecting PR1 and other loci not near the ELENA1 locus. However, the mechanism that gives specificity for these immunity-related loci remains to be determined.
IN BRIEF by Jennifer Mach jmach@aspb.org
REFERENCES
Chekanova, J.A., (2015) Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol 27, 207-216.
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., Arenas-Huertero, C., and Chua, N.H. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333-4345.
Seo, J.S., Sun, H.-X., Park, B.S., Huang, C.H., Yeh, S.-D., Jung, C., and Chua, N.-H. (2017) ELF18-INDUCED LONG NONCODING RNA associates with Mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 10.1105/tpc.16.00886.



Leave a Reply
Want to join the discussion?Feel free to contribute!